* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Translation
Expression vector wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Paracrine signalling wikipedia , lookup
Interactome wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
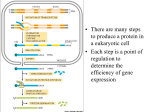
Gene expression wikipedia , lookup
Western blot wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein purification wikipedia , lookup
Genetic code wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Metalloprotein wikipedia , lookup
Messenger RNA wikipedia , lookup
Proteolysis wikipedia , lookup
Biosynthesis wikipedia , lookup