* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 6 Protein_Synthesis - bloodhounds Incorporated
Western blot wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Protein adsorption wikipedia , lookup
Molecular cloning wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular evolution wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
List of types of proteins wikipedia , lookup
Expanded genetic code wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemistry wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Non-coding RNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Genetic code wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene expression wikipedia , lookup
Messenger RNA wikipedia , lookup
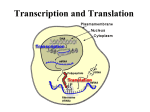
PHYSIOLOGY Signal Transduction and Protein Synthesis DNA DNA – Deoxyribonucleic Acid – Twisted ladder or double helix – Nucleotides » Composed of alternating sugar (Deoxyribose) and phosphate molecules and » Nitrogen bases Purines = adenine and guanine Pyrimidines = thymine cytosine DNA Purines bond with Pyrimidines – Complementary base pairs » Adenine with Thymine » Guanine with Cytosine DNA Purines bond with Pyrimidines – Complementary base pairs » Adenine with Thymine » Guanine with Cytosine Nucleoside – Sugar bonding with a base Nucleotide – Adding a phosphate to a nucleoside Phosphates attach to the 5’ carbon of the sugar Orientation of DNA The carbon atoms on the sugar ring are numbered for reference. The 5’ and 3’ hydroxyl groups (highlighted on the left) are used to attach phosphate groups. The directionality of a DNA strand is due to the orientation of the phosphate-sugar backbone. DNA is a double helix P T A P G 5’ 3’ P A sugar and phosphate “backbone” connects nucleotides in a chain. C DNA has directionality. PP P C G A P P Two nucleotide chains together wind into a helix. T P P G Hydrogen bonds between paired bases hold the two DNA strands together. C P P 3’ C G P DNA strands are antiparallel. 5’ DNA A chromosome – 23 pair = diploid – 23 = haploid; sex cells – Duplicating DNA structure tightly packed around histone proteins to form a nucleosome. DNA A gene – A series of bases that occupy a specific location (locus) on a chromosome – The code of a single protein or polypeptide Genetic Alphabet – Triplet = Three nucleotides on DNA with their corresponding base pairs making up the code of a single amino acid – Codon = Three successive nucleotides on RNA with their corresponding base pairs making up the code of a single amino acid – 20 amino acids – A series of amino acids makes up a protein DNA Consists of 3 billion base pairs – Codes for about 50 to 100,000 genes – Genes may exist in alternate forms = alleles » One allele from mom and one allele from dad – Nucleotide changes or mutations may occur in a gene » Sickle cell anemia – In a healthy population, a gene may exist in multiple alleles – Genetic Polymorphism = Multiple different forms at a gene locus in a population » Basis for DNA typing using MHC Terminology – Allele » An alternate form of a gene – Locus » Location of a gene on a chromosome – Gene » Genetic code or “blueprint” for the cell to build one particular protein http://www.youtube.com/watch?v=983lhh2 0rGY http://www.youtube.com/user/ndsuvirtualce ll?blend=10&ob=5 http://www.youtube.com/watch?v=YjWuVr zvZYA http://www.youtube.com/watch?v=FVuAw BGw_pQ&feature=related http://www.youtube.com/watch?v=5bLEDd -PSTQ&feature=related http://www.youtube.com/watch?v=NJxobgk PEAo Two types of nucleic acids DNA RNA Usually Has single-stranded uracil as a base Ribose as the sugar Carries protein-encoding information Can be catalytic Usually Has double-stranded thymine as a base Deoxyribose Carries as the sugar RNA-encoding information Not catalytic Protein Synthesis Proteins are necessary for cell functions Protein synthesis is under nuclear direction DNA specifies Proteins ? DNA mRNA ? Protein Redundancy of Genetic Code (p 115) A combination of three bases forms a codon 1 start codon 3 stop codon 60 other codons for 19 aa RNA Definitions – Exon » Amino acid specifying informational sequences in the genes of higher organisms – Intron » Noncoding segments or portions of DNA that ranges from 60 to 100,000 nucleotides long Transcription DNA is transcribed into complementary mRNA by RNA Polymerase + nucleotides + Mg2+ + ATP Gene = elementary unit of inheritance Compare to Fig. 4-33 http://vcell.ndsu.edu/animations/ Transcription First steps in protein synthesis that occurs completely within the nucleus DNA is used as a template to create a small single strand of mRNA that can leave through the nuclear pore. The enzyme RNA polymerase plus magnesium or manganese ions along with ATP are needed in this process. Transcription DNA is used as a template for creation of RNA using the enzyme RNA polymerase. DNA 5 ’ G T C A T T C G G 3’ 3’ C A G T A A G C C 5’ Transcription The new RNA molecule is formed by incorporating nucleotides that are complementary to the template strand. DNA coding strand 5 ’ DNA G T C A T T C G G 3’ 3’ G U C A U U C G G 3’ C A G T A A G C C 5’ DNA template strand 5 ’ RNA Transcription Promoter – Sequence on DNA where the RNA polymerase attaches to begin transcription – A region at the beginning of a gene that must be activated before transcription can begin. – This region is not transcribed into mRNA Transcription Transcription Factors – Binds to DNA and activates the promoter » Tells the RNA polymerase where to bind to the DNA » RNA polymerase moves along the DNA molecule and “unwinds” the double strand by breaking hydrogen bonds between base pairs – Sense strand » Guides RNA polymerase in RNA synthesis – Antisense strand » Sits idly by and is not transcribed Transcription Each base of the DNA sense strand pairs with a complementary mRNA base – AGTAC on DNA – UCAUG on mRNA Uracil is substituted for Thymine Ribose sugar is used as the backbone of mRNA instead of Deoxyribose sugar Initiation of transcription Transcription begins at the 5’ end of the gene in a region called the promoter. The promoter recruits TATA protein, a DNA binding protein, which in turn recruits other proteins. TATA binding protein Promoter DNA GG Transcription factor Gene sequence to be transcribed TATA CCC TATA box Transcription begins mRNA processing Alternative splicing occurs – Enzymes clip segments out of the middle or off the ends of mRNA strands » Introns – mRNA segments are spliced back together by the spliceozyme enzyme » Exons The processes mRNA leaves through the nuclear pore and attaches to a ribosome mRNA Contains the coded information for the amino acid sequence of a protein 3 main parts: – 5' leader sequence - important for the start of protein synthesis. – Coding Sequence - the sequence that codes for the amino acid. – 3’ trailer sequence - poly A tail. Messenger RNA undergoes three (or four) post-transcriptional modifications 1. Capping of 5’ end 2. Additional of poly A tail to 3’ end 3. Removal of introns 4. Editing of RNA (rarely) EUKARYOTES ONLY!!!!!!!!!!!!!!!! 5’ capping. Involves the addition of a guanine (usually 7-methylguanosine) to the terminal 5’ nucleotide. The enzyme that completes this process is called a capping enzyme. The 5’ cap is required for the ribosome to bind to the mRNA as the initial step of translation. Addition of a 3’poly A tail. This poly(A) tail is usually about 50 - 250 bps of adenine in length. There is no DNA template for this tail? Poly A tails are found on most mRNA molecules but not all (ex. histones mRNA have no poly A tail). In general, a eukaryotic mRNA molecule is longer than the required transcript. The enzyme RNA endonuclease cleaves the molecule at the poly(A) addition site to generate a 3’ OH end. The poly A tail is important for determining the stability of the mRNA molecule so the mRNA doesn’t degrade. Translation Translation begins when mRNA binds to a ribosome in the cytoplasm of the cell. Translation mRNA is translated into string of aa (= polypeptide) 2 important components ?? mRNA + ribosomes + tRNA meet in cytoplasm Anticodon pairs with mRNA codon aa determined Amino acids are linked via peptide bond. The Genetic Code The code has start and stop signals. AUG (methionine) is the common start codon Methionine can also be used WITHIN a polypeptide GUG may also be used as a start codon. There are three stop codons. UAG UAA UGA All three are chain termination codons. Ribosomal RNA Large and small subunits Binding sites – One for mRNA – Three for tRNA » P site = Peptidyl-tRNA site » A site = Aminoacyl-tRNA site » E site = Exit site Transfer RNA The correct amino acid is added to the growing polypeptide only if: – 1 - The appropriate amino acid is added to the tRNA by aminoacyl-tRNA synthetases. – 2 – Complementary binding occurs between the codon of the mRNA and the anticodon of the tRNA. Translation (An Overview) Translation is defined as protein synthesis. Occurs on ribosomes, where the genetic information is translated from the mRNA to a protein. mRNA is translated in the 5’ to 3’ direction. Amino acids are brought to the ribosome bound to a specific tRNA molecule. The mRNA and tRNA are responsible for the correct recognition of each amino acid in the growing polypeptide Initiation A small ribosomal subunit binds to both mRNA at the 5’ cap along with a specific initiator tRNA – The initiator tRNA carries methionine tRNA’s anticodon binds with the codon on mRNA The large ribosomal subunit attaches to form the translation initiation complex. The initiation complex is held together by proteins called initiation factors Initiation The tRNA sits in the P site of the ribosome The A site is vacant The methionine is at the N-terminus of the growing protein The carboxyl end is called the C-terminus All proteins grow from the N to the C-terminus Elongation Binding of the aminoacyl-tRNA to the ribosome formation of a peptide bond The movement (translocation) of the ribosome along the mRNA, one codon at a time. Elongation Three step cycle – The ribosome will move 5’ to 3’ on the mRNA Step one – The anticodon of an incoming aminoacyltRNA base-pairs with the complementary mRNA codon in the A site – GTP hydrolysis occurs Elongation Step two – The large ribosomal subunit catalyzes the formation of a peptide bond – Hydrogen bonds break between the t-RNA in the P site and between the codon and anti-codon Step three – translocation – The ribosomes moves along the mRNA one codon – The tRNA that was in the A site is now in the P site – The tRNA in the P site exits through a tunnel in the rRNA called the E site – The next tRNA enters in the empty A site Termination Termination is usually signaled by one of the three stop codons UAG, UAA or UGA. There are a number of “helper” proteins involved (e.g. termination factors and release factors). GTP is necessary to break the complex apart Translation initiation Leader sequence Small ribosomal subunit 5’ 3’ mRNA mRNA U U C G U C A U G G G A U G U A A G C G A A U A C Assembling to begin translation Initiator tRNA Met Translation Elongation Ribosome 5’ 3’ mRNA A U G G G A U G U A A G C G A U A C C C U tRNA P A Amino acid Met Gly Large ribosomal subunit Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A U A C C C U A P Met Gly Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A C C U A C A P Gly Cys A Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A P C C U A C A Gly Cys A Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A A C A U U C P Cys Lengthening polypeptide (amino acid chain) Lys A Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A A C A U U C P Cys Lys A Translation Elongation 5’ 3’ mRNA A U G G G A U G U A A G C G A A C A U U C P Cys Lys A Translation Elongation Stop codon 5’ mRNA A U G G G A U G U A A G C G A U A A U U C A P Lys Release factor Translation Termination Stop codon Ribosome reaches stop codon 5’ mRNA A U G G G A U G U A A G C G A U A A P Release factor A Translation Termination Once stop codon is reached, elements disassemble. Release factor P A Translation Modifications Protein folding Glycosylation – Addition of glycogen to the protein by the Golgi Apparatus – Create a glycoprotein Vesiculation – Protein is surrounded by a vesicle Exocytosis will then occur Post – Translational protein modifications: Folding, cleavage, additions glyco- , lipo- proteins Protein Sorting No signal sequence protein stays in cell Signal sequence protein destined for translocation into organelles or for export For “export proteins”: Signal sequence leads growing polypeptide chain across ER membrane into ER lumen Modifications in ER Transition vesicles to Golgi apparatus for further modifications Transport vesicles to cell membrane Signal Transduction 1st The signal molecule is a ligand that binds to a receptor. The ligand is also known as the first messenger because it brings information to its target cell 2nd Ligand-receptor binding activates the receptor 3rd The receptor in turn activates one or more intracellular signal molecules 4th the last signal molecule in the pathway initiates synthesis of target proteins or modifies existing target proteins to create a response Receptor Proteins Lipophilic signal molecules – Can diffuse through the phospholipid bilayer and bind to cytosolic receptors or nuclear receptors – Steroids are lipophilic Lipophobic signal molecules – Unable to diffuse through the phospholipid bilayer of the cell – Bind to receptor proteins on the cell membrane Receptor-Enzymes Transmembrane receptor binds with a ligand on the extracellular surface of the cell Intracellularly an enzyme is bound to the receptor protein – The enzyme is typically a protein kinase (ie. tyrosin kinase) or guanylyl cyclase – Guanylyl cyclase converts GTP to cyclic GMP (cGMP) – Adenylyl cyclase converts ATP to cyclic AMP (cAMP) Signal Transduction The process by which an extracellular signal molecule activates a membrane receptor that in turn alters intracellular molecules to create a response Signal Amplification Turns on signal molecules into multiple second messenger molecules Steps of Signal Transduction An extracellular signal molecule binds to and activates a protein or glycoprotein membrane receptor The activated membrane receptor turns on its associated proteins – The proteins may activate protein kinases – The proteins may create an intracellular second messenger Second Messenger Second messenger molecules – Alter the ion channels by opening or closing them – Increase intracellular calcium in order for the calcium to bind to proteins and change their function – Change enzyme activity Signal molecule binds to the G-protein linked receptors – The protein changes confirmation and activates the intracellular G protein The G protein moves horizontally in the membrane to bind with adenylyl cyclase, an amplifier enzyme Adenylyl cyclase converts ATP to cyclic AMP cAMP activates protein kinase A Protein kinase A phosphorylates other proteins – There is a cellular response » Such as a protein binding to the promoter site on DNA to start transcription » Release of calcium to change enzyme activity Specificity v Competition Receptors have binding sites for ligands – Different molecules may be able to bind to the same receptor » Ie. Epinephrine and its cousin Norepinephrine These both bind to a class of receptors called Adrenergic receptors – Alpha and Beta receptors » Alpha has a higher affinity for norepinephrine » B2 receptors have a higher affinity for epinephrine Agonists v Antagonists When a ligand combines with a receptor – Either the ligand turns the receptor on and elicits a response or – The ligand occupies the binding site and prevents a response from happening Agonist – turns receptors “on” Antagonist – turns receptors “off” Which of the following nucleotide bases in DNA can form H-bonds with the base adenine? A. Thymine B. Uracil C. Guanine D. Cytosine E. Both A and B Which of the following nitrogen bases are purines? A. Uracil and Guanine B. Adenine and Thymine C. Guanine and Cytosine D. Cytosine and Adenine E. Adenine and Guanine How many pair of chromosomes are found in a diploid cell? A. B. C. D. E. 8 16 23 46 0 DNA is formed by hydrogen bonding two antiparallel strands. A. B. True False In which direction is a DNA strand read? A. B. C. D. 5’ to 3’ 3’ to 5’ 3’ to 3’ 5’ to 5’ In DNA, guanine is bonded to cytosine by A. B. C. D. ionic bonding coordinate covalent bonding covalent bonding hydrogen bonding A gene can best be defined as: A. Three base triplet that specifies a particular amino acid B. Non-coding segments of DNA up to 100,000 nucleotides long. C. A segment of DNA that carries the instructions for one polypeptide chain. D. An RNA messenger that codes for a particular polypeptide. If the nucleotide or base sequence of the DNA strand used as a template for messenger RNA synthesis is ACGTT, then the sequence of bases in the corresponding mRNA would be: A. B. C. D. TGCAA ACGTT UGCAA GUACC In DNA, complementary base pairing occurs between _______________ A. B. C. D. Cytosine and thymine Adenine and guanine Thymine and uracil Guanine and cytosine Messenger RNA A. B. C. D. is composed of two nucleotide chains similar to DNA is a very stable molecule; that is, it is not easily broken down transfers genetic information from DNA molecules to the ribosome is synthesized on the ribosomes in the cytoplasm Transcription A. Occurs on the surface of the ribosome B. Is the final process in the assembly of a protein C. Occurs during the synthesis of any type of RNA by use of a DNA template D. Is catalyzed by DNA polymerase ___________ is an enzyme that breaks the hydrogen bonds on DNA to begin the process of transcription A. B. C. D. E. DNA polymerase RNA polymerase Lipase Phenylketonurinase Aldolase The genetic code on mRNA for methionine is A. B. C. D. AUG AUC UAG UAC Where on DNA does cAMP bind? A. B. C. D. Initiator site Promoter site DNA binding site TATA box