* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download LECTURE 18 - Budostuff
Magnesium in biology wikipedia , lookup
Biochemical cascade wikipedia , lookup
Butyric acid wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Phosphorylation wikipedia , lookup
Photosynthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Mitochondrion wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Microbial metabolism wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Electron transport chain wikipedia , lookup
Biochemistry wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
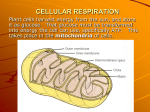
Summer School 2015 L6 – Cellular respiration Dr Agnieszka Adamczewska Images from Wikimedia Commons Major Concepts 1. Identify the three end-product options for glycolysis, and under what conditions these end-products form 2. State in words (not chemical formulae) the overall reaction of the glycolytic pathway, understand parts that are common and different 3. Understand how the overall balance sheet for glycolysis is obtained, and show the methods of reaction “coupling” that the cell uses 4. Say where the enzymes of the glycolytic pathway are to be found in eukaryotic cells and in prokaryotic cells 5. Explain what oxidation reduction reactions are and the special role of the coenzyme NAD+/NADH Major Concepts 6. 7. 8. 9. Give the names of, and recognise the equations for, the overall reactions of, the two major parts of the cellular respiration sequence - glycolysis and tricarboxylic acid (TCA) cycle. Describe how a H+ pumping mechanism is coupled to a proton-driven ATP synthase. State how many ATP molecules are produced per glucose molecule in the glycolytic pathway and in the whole respiratory pathway. Describe where in the respiratory pathway CO2 is released, and where O2 is consumed. Macromolecules store energy Energy is stored in carbon-carbon bonds e.g. glycogen or starch, fats and oils Get energy out of food through catabolic reactions Image from Campbell Biology 8e Australian Version © Pearson Education Inc. Metabolism (L3) Metabolism: chemical reactions that occur within cells • Catabolism – breaking down organic matter to release energy • Anabolism – using energy to produce cellular components Complex molecules Simple molecules Catabolism Pi ADP Anabolism ATP ATP: energy carrier Made through metabolism of energy rich molecules. - carbohydrates are converted into glucose - lipids are processed by β-oxidation Complex molecules Simple molecules Catabolism Pi ADP ATP ATP: Adenosine triphosphate Hydrolysis ADP Adenosine diphosphate Energy conversions in cells Energy from macromolecules is released by cellular respiration. Initial breakdown of macromolecules produces simple sugars, fatty acids, glycerol, and amino acids. Subsequent gradual oxidation of the fuel molecules by removal of electrons from C-C and C-H bonds releases energy: Energy conversions located in the cytosol 1. Glycolysis converts glucose to pyruvate 2. Fermentation to lactate and alcohol Energy conversions located in mitochondria in the presence of O2 3. -oxidation of lipids produces acetyl CoA 4. Citric acid cycle converts pyruvate to acetyl CoA and finally CO2 5. Electron transport chain (NADH, FADH2 > O2 > H2O) drives proton pumps, proton gradient is coupled to synthesis of ATP Energy conversion pathways Cellular respiration CYTOSOL 1 2 MITOCHONDRION 3 5 4 Glycolysis Pathway described in 1930's - a major biochemical triumph - involves a number of steps one glucose (C6) two pyruvate (2 x C3) Glycolysis Cytosol Glucose (6C) 2 ATP 3 steps 2 ADP Fructose 1,6-bisphosphate (unstable) G3P (3C) G3P (3C) NAD+ NADH 2 ADP 2 ATP 5 steps Pyruvate (3C) Mitochondria image from Wikimedia Commons glyceraldehyde 3 phosphate NAD+ NADH 2 ADP 2 ATP Pyruvate (3C) Glycolysis Energy Conversions Net yield 2 NADH 2 ATP Glycolysis “Splitting glucose” Glucose • from hydrolysis of polysaccharides (L2) • enters cell via facilitated diffusion - Glucose-Na+ symport (L5) Cytosol – 10 enzymes Glucose (6C) + 2 ATP + 2 NAD+ + 2 ADP + 2 Pi 2 Pyruvate (3C) + 4 ATP + 2 NADH Net yield: 2 ATP NAD+ Nicotinamide Adenine Dinucleotide NAD+ reduced to NADH by transfer of H+ from food Image from Wikimedia Commons Electron carrier (coenzyme ) 1. Glycolysis: Processing of glucose to pyruvate a) Glucose (6C) is phosphorylated using 2 ATP and split into two molecules of glyceraldehyde 3-phosphate (3C). Total 5 steps, consuming 2 ATP b) Oxidation in another 5 steps to 2 molecules of pyruvate (3C) and production of 4 ATP (net yield =2 ATP/glucose) • Pyruvate can be converted in the absence of O2 by alcoholic fermentation to ethanol (in yeast and bacteria) or by lactate fermentation to lactate (muscle tissue) • In the presence of O2 pyruvate enters mitochondria, is converted (decarboxylated) to a 2C compound acetyl CoA (a substrate for citric acid cycle) CO2, and NADH; 2. Fermentation Anaerobic conversion of pyruvate to alcohol or lactic acid Alcohol fermentation By yeast and many bacteria. Used by humans for thousands of years in brewing, winemaking, and baking (CO2 bubbles from baker’s yeast) REDUCTION Lactic acid fermentation By certain fungi and bacteria Used in dairy industry to make cheese and yogurt A product of muscle exercise, may enhance muscle performance (pain from K+ ions) REDUCTION Fermentation No oxygen 2ADP + 2Pi 1× Glucose Glycolysis then fermentation Glycolysis 2NAD+ Fungi, bacteria, animals 2ATP 2× Pyruvate 2NADH + 2H+ 2× Lactic acid lactic acid 2ADP + 2Pi Yeast, plants 1× Glucose ethanol + CO2 Glycolysis 2NAD+ 2× Ethanol 2ATP 2× Pyruvate 2NADH + 2H+ 2× CO2 Pyruvate A key juncture in catabolism Food Chloroplast Mitochondria (L4) • Site of cellular respiration (energy production) • Double membrane: permeable outer membrane and impermeable, folded inner membrane (cristae) containing enzymes of respiration Inner membrane Outer membrane Matrix Crista Images from Wikimedia Commons Mitochondria: number, shape, and subcellular location are highly variable Fuel in, energy out Glucose + oxygen carbon dioxide + water + energy Cytosol NADH FADH2 Oxidative phosphorylation Krebs cycle NADH Glycolysis Crista Matrix ATP Mitochondria image from Wikimedia Commons Intermembrane space Mitochondria ATP ATP Intermediate reaction Pyruvate to Acetyl CoA Outer mitochondrial membrane Inner mitochondrial membrane CoA Matrix CO2 Acetyl-CoA CoA Pyruvate NADH + H+ NAD+ Mitochondria image from Wikimedia Commons Pyruvate dehydrogenase complex Other fuel molecules • Other carbohydrates (apart from glucose) • Fats and proteins can also be broken down and enter pathways Image from Campbell Biology 8e Australian Version © Pearson Education Inc. -oxidation 3. -Oxidation of lipids -oxidation degrades long-chain fatty acids by 2C atoms at a time Last reaction splits off acetyl CoA (energy in C-C bond) and enters Citric acid cycle NADH, FADH2, energy (electron) carriers h 4. Krebs cycle 8 steps, each catalysed by different enzyme TCA cycle Citric acid cycle AcetylCoA (2C) Oxaloacetate (4C) 4C Citrate (6C) NADH 6C Krebs cycle 4C FADH2 NADH CO2 NAD+ and FAD reduced to NADH and FADH2 accept e- from intermediates NADH 5C 4C 4C ATP CO2 CO2 evolved ATP is formed Chemical bonds to electrons C6H12O6 + 0O2 6CO2 + 0H2O + energy 6× NADH glucose CO2 2× NADH 2× FADH2 Krebs cycle Glycolysis ATP Mitochondria image from Wikimedia Commons ATP CO2 Chemical bonds to electrons 1 glucose: Glycolysis 2 pyruvate + 2 ATP + 2 NADH 2 pyruvate 2 acetyl CoA + 2 NADH + 2 CO2 TCA cycle: 2 acetyl CoA 6 NADH + 2 FADH2 + 4 CO2 + 2 ATP 1 glucose 4 ATP + 8 NADH + 2 FADH2 + 6 CO2 5. Electron transport chain (ETC) (Proton-motive force i.e. the power in movement of protons) NADH and FADH2 Inner membrane of mitochondria Inner membrane Four protein complexes of acceptors Oxygen needed Matrix Crista Mitochondria image from Wikimedia Commons 5. Electron transport chain Start with NADH (or FADH2) as primary electron donor Intermembrane space H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ Inner mitochondrial membrane C Finish with O2 as terminal electron acceptor Q e– e– H+ NADH FADH2 NAD+ NADH dehydrogenase Mitochondrial matrix H+ 2H+ + 1/2O2 FAD bc1 complex H+ H2O Cytochrome oxidase complex Chain of redox reactions Inner mitochondrial membrane Electrons move to higher redox potentials, towards oxygen with highest electron affinity Energy released is used to pump H+ from matrix to intermembrane space NADH + H+ Energy released 2H+ + NAD+ 2e High energy Electrons “falling” from NADH to oxygen Cyt b (Fe3+) Cyt b (Fe2+) Cyt c (Fe2+) Cyt c (Fe3+) Cyt a (Fe3+) Cyt a (Fe2+) H 2O Low energy ½ O2 + 2H+ Generating proton gradient Start with NADH (or FADH2) as primary electron donor Intermembrane space H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ H+ Inner mitochondrial membrane C Finish with O2 as terminal electron acceptor Q e– e– H+ NADH FADH2 NAD+ NADH dehydrogenase Mitochondrial matrix H+ 2H+ + 1/2O2 FAD bc1 complex H+ H2O Cytochrome oxidase complex Chemiosmosis couples the ETC to ATP synthesis! OMM IMS ATP synthase IMM FADH2 FAD NADH + H+ NAD+ H 2O ½ O2 + 2H+ H+ + OH- Electron transport chain H 2O ADP + Pi ATP ATP synthase complexity of 1o, 2o, 3o and 4o structure Intermembrane space Inner membrane embedded in inner membrane of mitochondria highly conserved in nature Matrix Image from http://www.rcsb.org/pdb/101/motm.do?momID=72 H+ ATP synthase H+ binding Rotor H+ ions flow down gradient and enter half channel in stator H+ H+ Intermembrane space Inner membrane H+ ions enter binding sites in rotor, changing shape of subunits so rotor spins Each H+ ion makes one complete turn before being released to matrix Catalytic knob Matrix Spinning of rotor causes internal rod to spin Turning rod activates catalytic sites in knob H+ ADP + Pi ATP Image from http://www.rcsb.org/pdb/101/motm.do?momID=72 Fuel in, energy out Glucose + oxygen carbon dioxide + water + energy Cytosol NADH FADH22 Oxidative phosphorylation Krebs cycle NADH Glycolysis Crista Matrix 2x ATP Mitochondria image from Wikimedia Commons Intermembrane space 2x Mitochondria ATP ~34x ATP Aerobic respiration = lots of energy C6H12O6 + 6 O2 6 CO2 + 6 H2O + ~38 ATP Where are these ATPs from? Glycolysis = 2 ATP TCA = 2 ATP Oxidative phosphorylation (ETC + chemiosmosis) = ~34 ATP Cristae increases surface area • Infoldings of the inner mitochondrial membrane (cristae) greatly increase the number of electron transport chain proteins and ATP synthase proteins Lack of oxygen? Need oxygen to accept final electrons If no oxygen, complex IV keeps electrons All protein complexes keep electrons No pumping of H+ into intermembrane space No H+ gradient No energy for ATP synthase No ATP made in ETC - ATP from glycolysis and TCA not enough Most cells cannot survive long without oxygen Prokaryotic cell – no mitochondria Still need ATP! Plasma membrane Capsule Cell wall Nucleoid region Flagellum Pilus Image from Wikimedia Commons Ribosome Aerobic Bacteria do it too! Enzymes embedded in bacterial cell membrane H+ ATP synthase complex ATP ADP + Pi Cell membrane O2 + H2O NAD NADH Electron carrier H+ Anaerobic respiration Prokaryotes living in anaerobic environment - no oxygen e.g. waterlogged soils, intestines Nitrate (NO3-) or sulfate (SO42-) are the terminal electron acceptors Products include CO2, inorganic substance, ATP e.g. C6H12O6 + 12 KNO3 (potassium nitrate) 6 CO2 + 6 H2O + 12 KNO3 (potassium nitrite) + ATP Summary Cytoplasm and mitochondria are sites of cellular respiration Glucose + oxygen carbon dioxide + water + energy Aerobic respiration has four stages For one glucose molecule: Glycolysis = 2 ATP, TCA = 2 ATP, Oxidative phosphorylation (ETC, chemiosmosis) = ~34 ATP Chemiosmosis - electrons in ETC used to pump protons into inter-membrane space, this establishes a proton gradient across inner membrane, protons accumulate, lowers pH Redox reactions integral to ETC Fermentation and anaerobic respiration in absence of O2 Aerobic respiration Stage Where Main starting Main end materials products Glycolysis Cytoplasm Glucose Pyruvate, ATP, NADH forming Acetyl CoA Matrix of mitochondria Pyruvate Acetyl CoA, CO2, NADH Citric acid cycle Matrix of mitochondria Acetyl CoA, H2O CO2, NADH, FADH2, ATP O2, NADH, FADH2 ATP, H2O Inner Oxidative membrane phosphorylation mitochondria • Read: • Knox B, et al. (2010) Biology: an Australian perspective. - Chapter 6 Harvesting energy