* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Inquiry into Life Twelfth Edition
Biochemistry wikipedia , lookup
Ligand binding assay wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
RNA interference wikipedia , lookup
Western blot wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Proteolysis wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein structure prediction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Genetic code wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene expression wikipedia , lookup
Biosynthesis wikipedia , lookup
Transfer RNA wikipedia , lookup
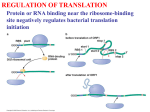
Lecture PowerPoint to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 17 The Mechanism of Translation I: Initiation Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. 17.1 Initiation of Translation in Bacteria • Two important events must occur before translation initiation can take place – Generate a supply of aminoacyl-tRNAs • Amino acids must be covalently bound to tRNAs • Process of bonding tRNA to amino acid is called tRNA charging – Dissociation of ribosomes into their two subunits • The cell assembles the initiation complex on the small ribosomal subunit • The two subunits must separate to make assembly possible 17-2 tRNA Charging • All tRNAs have same 3 bases at 3’-end (CCA) • Terminal adenosine is the target for charging with amino acid • Amino acid attached by ester bond between – Its carboxyl group – 2’- or 3’-hydroxyl group of terminal adenosine of tRNA 17-3 Two-Step Charging • Aminoacyl-tRNA synthetases join amino acids to their cognate tRNAs • This is done in a two-step reaction: – Begins with activation of the amino acid with AMP derived from ATP – In the second step, the energy from the aminoacyl-AMP is used to transfer the amino acid to the tRNA 17-4 Aminoacyl-tRNA Synthetase Activity 17-5 Dissociation of Ribosomes • E. coli ribosomes dissociate into subunits at the end of each round of translation • IF1 actively promotes this dissociation • IF3 binds to free 30S subunit and prevents reassociation with 50S subunit to form a whole ribosome 17-6 Ribosomal Subunit Exchange 17-7 Formation of the 30S Initiation Complex When ribosomes have been dissociated into 50S and 30S subunits, cell builds a complex on the 30S subunit: – mRNA – Aminoacyl-tRNA – Initiation factors • IF3 binds by itself to 30S subunit • IF1 and IF2 stabilize this binding • IF2 can bind alone, but is stabilized with help of IF1 and IF3 • IF1 does not bind alone 17-8 First Codon and the First Aminoacyl-tRNA • Prokaryotic initiation codon is: – Usually AUG – Can be GUG – Rarely UUG • Initiating aminoacyl-tRNA is N-formylmethionyl-tRNA • N-formyl-methionine (fMet) is the first amino acid incorporated into a polypeptide • This amino acid is frequently removed from the protein during maturation 17-9 N-Formyl-Methionine 17-10 Binding mRNA to the 30S Ribosomal Subunit • The 30S initiation complex is formed from a free 30S ribosomal subunit plus mRNA and fMet-tRNA • Binding between the 30S prokaryotic ribosomal subunit and the initiation site of a message depends on base pairing between – Short RNA sequence • Shine-Dalgarno sequence • Upstream of initiation codon – Complementary sequence • 3’-end of 16S RNA 17-11 Initiation Factors and 30S Subunit • Binding of the Shine-Dalgarno sequence with the complementary sequence of the 16S rRNA is mediated by IF3 – Assisted by IF1 and IF2 – All 3 initiation factors have bound to the 30S subunit at this time 17-12 Binding of fMet-tRNA to the 30S Initiation Complex • IF2 is the major factor promoting binding of fMet-tRNA to the 30S initiation complex • Two other initiation factors also play an important supporting role • GTP is also required for IF2 binding at physiological IF2 concentrations • The GTP is not hydrolyzed in the process 17-13 30S Initiation Complex The complete 30S initiation complex contains one each: – 30S ribosomal subunit – mRNA – fMet-tRNA – GTP – Factors IF1, IF2, IF3 17-14 Formation of the 70S Initiation Complex • GTP is hydrolyzed after the 50S subunit joins the 30S complex to form the 70S initiation complex • This GTP hydrolysis is carried out by IF2 in conjunction with the 50S ribosomal subunit • Hydrolysis purpose is to release IF2 and GTP from the complex so polypeptide chain elongation can begin 17-15 Bacterial Translation Initiation 1. IF1 influences dissociation of 70S ribosome to 50S and 30S 2. Binding IF3 to 30S, prevents subunit reassociation 3. IF1, IF2, GTP bind alongside IF3 4. Binding mRNA to fMet-tRNA forming 30S initiation complex a. Can bind in either order b. IF2 sponsors fMet-tRNA c. IF3 sponsors mRNA 5. Binding of 50S with loss of IF1 and IF3 6. IF2 dissociation and GTP hydrolysis 17-16 17.2 Initiation in Eukaryotes •Eukaryotic – Begins with methionine – Initiating tRNA not same as tRNA for interior – No Shine-Dalgarno – mRNA have caps at 5’end • Bacterial – N-formyl-methionine – Shine-Dalgarno sequence to show ribosomes where to start 17-17 Scanning Model of Initiation • Eukaryotic 40S ribosomal subunits locate start codon by binding to 5’-cap and scanning downstream to find the 1st AUG in a favorable context • Kozak’s Rules are a set of requirements • Best context uses A of ACCAUGG as +1: – Purine in -3 position – G in +4 position • 5-10% cases ribosomal subunits bypass 1st AUG scanning for more favorable one 17-18 Translation With a Short ORF • Ribosomes can use a short upstream open reading frame: – Initiate at an upstream AUG – Translate a short Open Reading Frame – Continue scanning – Reinitiate at a downstream AUG 17-19 Scanning Model for Translation Initiation 17-20 Effects of mRNA Secondary Structure • Secondary structure near the 5’-end of an mRNA can have either positive or negative effects • Hairpin just past an AUG can force a pause by ribosomal subunit and stimulate translation • Very stable stem loop between cap and initiation site can block scanning and inhibit translation 17-21 Eukaryotic Initiation Factors • Bacterial translation initiation requires initiation factors as does eukaryotic initiation of translation • Eukaryotic system is more complex than bacterial – Scanning process – Factors to recognize the 5’-end cap 17-22 Translation Initiation in Eukaryotes Eukaryotic initiation factors and general functions: • eIF2 binds Met-tRNA to ribosomes • eIF2B activates eIF2 replacing its GDP with GTP • eIF1 and eIF1A aid in scanning to initiation codon • eIF3 binds to 40S ribosomal subunit, inhibits reassociation with 60S subunit • eIF4 is a cap-binding protein allowing 40S subunit to bind 5’-end of mRNA • eIF5 encourages association between 60S ribosome subunit and 48S complex • eIF6 binds to 60S subunit, blocks reassociation with 40S subunit 17-23 Function of eIF4 • eIF4 is a cap-binding protein • This protein is composed of 3 parts: – eIF4E, 24-kD, has actual cap binding activity – eIF4A, a 50-kD polypeptide – eIF4G is a 220-kD polypeptide • The complex of the three polypeptides together is called eIF4F 17-24 Function of eIF4A and eIF4B • eIF4A – Has an RNA helicase activity – This activity unwinds hairpins found in the 5’leaders of eukaryotic mRNA – Unwinding activity is ATP dependent • eIF4B – Has an RNA-binding domain – Can stimulate the binding of eIF4A to mRNA 17-25 Function of eIF4G • eIF4G is an adaptor protein capable of binding to other proteins including: – eIF4E, cap-binding protein – eIF3, 40S ribosomal subunit-binding protein – Pab1p, a poly[A]-binding protein • Interacting with these proteins lets eIF4G recruit 40S ribosomal subunits to mRNA and stimulate translation 17-26 Structure and Function of eIF3 • eIF3 is a 5-lobed protein that binds at the same site to: – eIF4G – Prominent part of viral IRES • This explains how the IRES can substitute for 40S ribosomal subunit to mRNA • Cryo-EM studies have produced a model for the eIF3-IRES-40S complex explaining how eIF3 prevents premature 40S-60S association 17-27 Prevention of Premature 40S60S Association • eIF3 blocks key contact point between subunits 40S and 60S • eIF4G, so also eIF4E, locate close to the cap on an mRNA bound to 40S ribosomal particle • eIF4 would be in position to cap-bind 17-28 Functions of eIF1 and eIF1A • eIF1 and eIF1A act synergistically to promote formation of a stable 48S complex involving: – Initiation factors – Met-tRNA – 40S ribosomal subunits bound at initiation codon of mRNA • eIF1 and eIF1A act by – Dissociating improper complexes between 40S subunits and mRNA – Encouraging formation of stable 48S complexes 17-29 Principle of the Toeprint Assay Source: Adapted from Jackson, R., J. G. Sliciano, Cinderella factors have a ball, Nature 394:830, 1998. 17-30 Functions of eIF5 and eIF5B • eIF5B is homologous to prokaryotic factor IF2 – Binds GTP • Uses GTP hydrolysis to promote its own dissociation from ribosome • Permits protein synthesis to begin – Stimulates association of 2 ribosomal subunits • Differs from IF2 as eIF5B cannot stimulate binding of initiating aminoacyl-tRNA to small ribosomal subunit • eIF5B works with eIF5 17-31 17.3 Control of Initiation • Given the amount of control at the transcriptional and posttranscriptional levels, why control gene expression at translational level? • Major advantage = speed – New gene products can be produced quickly – Simply turn on translation of preexisting mRNA • Valuable in eukaryotes • Transcripts are relatively long • Take correspondingly long time to make – Most control of translation happens at the initiation step 17-32 Bacterial Translational Control • Most bacterial gene expression is controlled at transcription level • Majority of bacterial mRNA has a very short lifetime – Only 1 to 3 minutes – Allows bacteria to respond quickly to changing circumstances • Different cistrons on a polycistronic transcript can be translated better than others 17-33 Shifts in mRNA Secondary Structure • mRNA secondary structure can govern translation initiation – Replicase gene of the MS2 class of phages • Initiation codon is buried in secondary structure until ribosomes translating the coat gene open up the structure – Heat shock sigma factor, s32 of E. coli • Repressed by secondary structure that is relaxed by heating • Heat can cause an immediate unmasking of initiation codons and burst of synthesis 17-34 Proteins/mRNAs Induce mRNA Secondary Structure Shifts • Small RNAs with proteins can affect mRNA 2° structure to control translation initiation • Riboswitches can be used to control translation initiation via mRNA 2° structure – 5’-untranslated region of E. coli thiM mRNA contain a riboswitch – This includes an aptamer that binds thiamine and its metabolite • Thiamine phosphate • Thiamine pyrophosphate (TTP) 17-35 Activation of mRNA Translation • When TPP abundant – Binds aptamer – Causes conformational shift in mRNA – Ties up Shine-Dalgarno in 2° structure • Shift hides the SD sequence from ribosomes – Inhibits translation of mRNA • Saves energy as thiM mRNA encodes an enzyme needed to produce more thiamine and TPP 17-36 Eukaryotic Translational Control • Eukaryotic mRNA lifetimes are relatively long, so there is more opportunity for translation control than in bacteria • eIF2 a-subunit is a favorite target for translation control – Heme-starved reticulocytes activate HCR (hemecontrolled repressor) • Phosphorylates eIF2a • Inhibit initiation – Virus-infected cells have another kinase, DAI – Phosphorylates eIF2a – Inhibits translation initiation 17-37 Phosphorylation of an eIF4EBinding Protein • Insulin and a number of growth factors stimulate a pathway involving a protein kinase known as mTOR • mTOR kinase’s target protein – PHAS-1 (rat) – 4E-BP1 (human) • Once phosphorylated by mTOR – This protein dissociates from eIF4E – Releases it to participate in active translation initiation 17-38 Repression of Translation by Phosphorylation 17-39 Stimulation of Translation by Phosphorylation of PHAS-1 17-40 Control of Translation Initiation by Maskin • In Xenopus oocytes, Maskin binds to eIF4E and to CPEB (cytoplasmic polyadenylation element binding-protein) • Maskin bound to eIF4E, cannot bind to eIF4G, translation is now inhibited • Upon activation of oocytes – CPEB is phosphorylated – Polyadenylation is stimulate – Maskin dissociates from eIF4E • When Maskin is no longer attached – eIF4E able to associate with eIF4G – Translation can initiate 17-41 Stimulation by an mRNABinding Protein • Ferritin mRNA translation is subject to induction by iron • Induction seems to work as follows: – Repressor protein (aconitase apoprotein) binds to stem loop iron response element (IRE) – Binding occurs near 5’-end of the 5’-UTR of the ferritin mRNA – Iron removes this repressor and allows mRNA translation to proceed 17-42 Blockage of Translation Initiation by an miRNA • The let-7 miRNA shifts the polysomal profile of target mRNAs in human cells toward smaller polysomes – This miRNA blocks translation initiation in human cells • Translation initiation that is cap-independent due to presence of an IRES, or a tethered initiation factor, is not affected by let-7 miRNA – This miRNA blocks binding of eIF4E to the cap of target mRNAs in the human cell 17-43