* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lect20.ProteinSynthesis
Bimolecular fluorescence complementation wikipedia , lookup
Protein folding wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Homology modeling wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
RNA-binding protein wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
Alpha helix wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
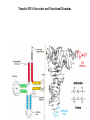
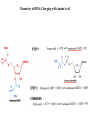
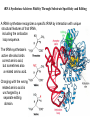
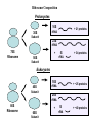
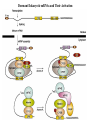
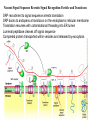
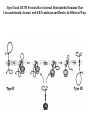
LECT 20: PROTEIN SYNTHESIS AND TRANSLATIONAL CONTROL High fidelity of protein synthesis from mRNA is essential. Mechanisms controling translation accuracy include very high fidelity of (1) tRNA charging with correct amino acid (2) codon-directed delivery of correct charged tRNA to ribosome Mechanisms for translation elongation and termination are very similar in prokaryotes and eukaryotes, but mechanisms of initiation are fundamentally different Translation of eukaryotic mRNAs can be regulated at the step of initiation by multiple mechanisms Secreted and membrane-spanning proteins are co-translationally delivered across membranes by “translocons” guided by signal and anchor sequences in the nascent polypeptide chain Transfer RNA Structure and Functional Domains Chemistry of tRNA Charging with Amino Acid tRNA Synthetase Achieves Fidelity Through Substrate Specificity and Editing A tRNA synthetase recognizes a specific tRNA by interaction with unique structural features of that tRNA, including the anticodon loop sequence. The tRNA synthetase’s active site also binds correct amino acid, but sometimes also a related amino acid. Charging with the wrong related amino acid is uncharged by a separate editing domain. Ribosome Composition Prokaryotes 16S 30S + 21 proteins rRNA Subunit 23S rRNA 70S Ribosome + 5S + 34 proteins rRNA 50S Subunit Eukaryotes 18S 40S + ~25 proteins rRNA Subunit 28S rRNA 80S Ribosome + 60S Subunit 5S rRNA + ~40 proteins Translation Initiation Complex in Prokaryotes Initiation factors deliver initiator fMet-tRNA to mRNA initiation codon positioned at the “P” site of the 30S ribosomal subunit. Initiation codon is AUG preceded by “Shine-Delgarno” sequence that is recognized by the 16S rRNA in the 30S ribosomal subunit. Translation Initiation Complex in Eukaryotes Initiation factors deliver 40S ribosomal subunit and Met-tRNA to 5’ methylated cap on mRNA. The 40S subunit then “scans” down mRNA and docks at first AUG codon. Initiation factor interaction with 5’ cap is facilitated by prior recruitment of initiation factor to the mRNA via polyA binding protein (PABP) bound to the mRNA’s 3’ polyA tail. Dormant Eukaryotic mRNAs and Their Activation Elongation Sequentially Adds Amino Acids to a Peptidyl-Acyl tRNA Peptide(n)-acyl-tRNA + Aminoacyl-tRNA ---> tRNA(uncharged) + Peptide(n + 1)-acyl-tRNA Elongation Factors and GTP Needed for tRNA Recruitment and Translocation EF-Tu GDP + Pi GTP Release Factors Dock Recognize Stop Codons at A Site To Promote Termination Upon release factor docking to stop codon, water is used instead of amino group of charged tRNA as nucleophile to attack acyl bond of peptidyl-acyl-tRNA. Hydrolysis releases peptide from tRNA and ribosome. Secreted Proteins Have N-Terminal Signal Sequence Signal sequence is 10-15 residue hydrophobic stretch near N-terminus. Signal sequence triggers secretion mechanism, is usually cleaved at nearby downstream small amino acid (Gly-X, Ser-X, Ala-X) Nascent Signal Sequence Recruits Signal Recognition Particle and Translocon SRP recruitment to signal sequence arrests translation SRP docks to andopens a translocon on the endoplasmic reticular membrane Translation resumes with cotranslational threading into ER lumen Lumenal peptidase cleaves off signal sequence Completed protein transported within vesicles and released by exocytosis Transmembrane Proteins Are Cotranslationally Inserted Into Membrane Type I TM protein has signal sequence and downstream hydrophobic helical transmembrane sequence and polar stop transfer element. Type II and III TM proteins do not have signal sequence. Type II and III TM Proteins Have Internal Hydrophobic Domains That Cotranslationally Interact with ER Translocon and Resolve In Different Ways