* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Phosphorylation wikipedia , lookup
Endomembrane system wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Signal transduction wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein phosphorylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Protein moonlighting wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein folding wikipedia , lookup
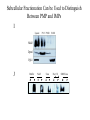
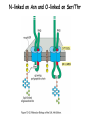
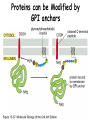
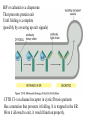
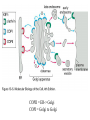
Lecture 2 Protein Glycosylation and Vesicular Sorting Subcellular Fractionation Can be Used to Distinguish Between PMP and IMPs I Lysate P13 P100 S100 Msb2 Dpm1 Pgk1 J Buffer NaCl Urea Na CO SDS/Urea Density Gradient Centrifugation Can be Used to Distinguish Between PMP and IMPs Figure 12-37b Molecular Biology of the Cell (© Garland Science 2008) Protein modification by Glycosylation The term "CDG-Syndrome“ stands for Congenital Disorders of Glycosylation, and describes an inherited metabolic disorder in which protein-glycosylation – the synthesis of carbohydrate-protein complexes – is impaired. Protein-glycosylation is a vital process, which plays an important role in a large number of biochemical and cellular processes. Cystic Fibrosis Emphysema Proteins are Glycosylated in ER and Golgi -important for folding -important for function -important for targeting N-linked on Asn and O-linked on Ser/Thr Biosynthesis of the Glycosyl Moiety Tunicamycin Streptomyces compound that blocks transfer of GlcNAc-1-P from UDPGlcNAc to dolichyl-P Two ways to show that a protein is glycosylated Tunicamycin - ADDED TO CELLS Endo H - a glycosidase that cleaves high mannose and some hybrid N-linked oligosaccharosides ADDED TO PURIFIED PROTEIN Sialic Acid glycosylation of Different Proteins Proteins can be Modified by GPI anchors BiP or calnexin is a chaperone That prevents protein exit Until folding is complete (possibly by covering up exit signals) CFTR Cl- ion channel receptor in cystic fibrosis patients Has a mutation that prevents it folding. It is trapped in the ER. Were it allowed to exit, it would function properly. Protein Folding by Chaperones Model of calnexin action. As a nascent polypeptide chain enters the ER, certain Asn residues are glycosylated through the addition of an oligosaccharide of composition Glc3Man9GlcNAc2. The outermost two glucoses are rapidly removed through the action of glucosidases I and II to reveal the monoglucosylated species recognized by the lectin sites of calnexin/calreticulin. In their ATP-bound state, calnexin bind to the monoglucosylated oligosaccharide and hydrophobic segments of the unfolded glycoprotein (via their polypeptide binding or chaperone sites). Glycoprotein dissociation involves not only the action of glucosidase II to remove the terminal glucose residue but also a change in affinity of the polypeptide binding site, possibly regulated by a shift from an ATP-bound to an ADP-bound or unbound state. After dissociation, if folding does not occur rapidly, the glycoprotein is reglucosylated by another ER enzyme, UDP-glucose:glycoprotein glucosyltransferase. This enzyme only reglucosylates non-native protein conformers. The glycoprotein can then re-bind in dual fashion to the ATP form of calnexin/calreticulin. In this model, both the glucosyltransferase and calnexin/calreticulin act as folding sensors. The function of this binding and release cycle is three-fold: 1) the polypeptide binding (chaperone) site prevents glycoprotein aggregation, 2) the lectin and polypeptide sites retain non-native conformers in the ER until a native structure is acquired (quality control), and 3) calnexin and calreticulin bring ERp57 into proximity with the non-native glycoprotein. ERp57 catalyzes disulfide bond formation and isomerization within the glycoprotein. Protein Misfolding leads to Aggregation and Disease (prions and Altzheimer’s) Unfolded Protein Response Taking out the Trash Unfolded Protein Response Ire1p is a transmembrane serine-threonine kinase, oriented with the amino terminus (N) in the ER lumen and the carboxyl terminus in the cytosol. When unfolded proteins accumulate in the ER, Ire1p oligomerizes, trans-autophosphorylates via the cytosolic kinase domain (K) and activates the endonuclease in the tail domain (T). The endonuclease Ire1p cuts HAC1 mRNA at two sites, removing a nonclassical intron; the two exons are rejoined by Rlg1p (tRNA ligase). HAC1u (‘uninduced’) is not translated owing to the presence of the intron, and Hac1pu is not produced (brackets). After Ire1-mediated splicing, HAC1i mRNA is efficiently translated into Hac1pi, a transcriptional activator that upregulates expression of UPR target genes after binding to the unfolded protein response element (UPRE) in the promoters of genes encoding ER-resident chaperones and other proteins. Vesicular Trafficking allows Proteins and Vesicles to Reach their Destinations QuickTime™ and a Sorenson Video 3 decompressor are needed to see this picture. The orientation of transmembrane proteins Proteins IN the ER face the OUTSIDE of the cell QuickTime™ and a MPEG-4 Video decompressor are needed to see this picture. QuickTime™ and a MPEG-4 Video decompressor are needed to see this picture. COPII = ER-> Golgi COPI = Golgi to Golgi