* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Gene knockout
Epigenetics of human development wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Genome evolution wikipedia , lookup
Public health genomics wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Oncogenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Point mutation wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome (book) wikipedia , lookup
Gene desert wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Helitron (biology) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Genome editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
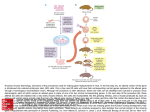
Gene knockout A gene knockout is a genetically engineered organism that carries one or more genes in its chromosomes that have been made inoperative (have been "knocked out" of the organism). This is done for research purposes. Also known as knockout organisms or simply knockouts, they are used in learning about a gene that has been sequenced, but which has an unknown or incompletely known function. Researchers draw inferences from the difference between the knockout organism and normal individuals. The term also refers to the process of creating such an organism, as in "knocking out" a gene. Knockout is accomplished through a combination of techniques, beginning in the test tube with a plasmid, a bacterial artificial chromosome or other DNA construct, and proceeding to cell culture. Individual cells are genetically transformed with the construct and--for knockouts in multi-cellular organisms--ultimately fused with a stem cell from a nascent embryo. The construct is engineered to recombine with the target gene, which is accomplished by incorporating sequences from the gene itself into the construct. Recombination then occurs in the region of that sequence within the gene, resulting in the insertion of a foreign sequence to disrupt the gene. With its sequence interrupted, the altered gene in most cases will be translated into a nonfunctional protein, if it is translated at all. A conditional knockout allows gene deletion in a tissue specific manner. Because recombination is a rare event in the case of most cells and most constructs, the foreign sequence chosen for insertion usually is a reporter. This enables easy selection of cells or individuals in which knockout was successful. In diploid organisms, which contain two alleles for most genes, and may as well contain several related genes that collaborate in the same role, additional rounds of transformation and selection are performed until every targeted gene is knocked out. Knock-in is similar to knock-out, but instead it replaces a gene with another instead of deleting it. Knockout mouse A knockout mouse is a genetically engineered mouse one or more of whose genes have been made inoperable through a gene knockout. Knockout is a route to learning about a gene that has been sequenced but has an unknown or incompletely known function. Mice are the laboratory animal species most closely related to humans in which the knockout technique can be easily performed, so they are a favourite subject for knockout experiments, especially with regard to genetic questions that relate to human physiology. (Gene knockout in rats is much harder and has only been possible since 2003.) Use Knocking out the activity of a gene provides information about what that gene normally does. Humans share many genes with mice. Consequently, observing the characteristics of knockout mice Create PDF with GO2PDF for free, if you wish to remove this line, click here to buy Virtual PDF Printer gives researchers information that can be used to better understand how a similar gene may cause or contribute to disease in humans. Examples of research in which knockout mice have been useful include studying and modelling different kinds of cancer, obesity, heart disease, diabetes, arthritis, substance abuse, anxiety, aging and Parkinson disease. Knockout mice also offer a biological context in which drugs and other therapies can be developed and tested. Many of these mouse models are named after the gene that has been inactivated. For example, the p53 knockout mouse is named after the p53 gene which codes for a protein that normally suppresses the growth of tumours by arresting cell division. Humans born with mutations that inactivate the p53 gene suffer from Li-Fraumeni syndrome, a condition that dramatically increases the risk of developing bone cancers, breast cancer and blood cancers at an early age. Other mouse models are named, often with creative flair, according to their physical characteristics or behaviours. For example, "Methuselah" is a knockout mouse model noted for longevity, while "Frantic" is a model useful for studying anxiety disorders. Procedure There are several variations to the procedure of producing knockout mice; the following is a typical example. 1. The gene to be knocked out is isolated from a mouse gene library. Then a new DNA sequence is engineered which is very similar to the original gene and its immediate neighbor sequence, except that it is changed sufficiently to make it inoperable. Usually, the new sequence is also given a marker gene, a gene that normal mice don't have and that transfers resistance to a certain antibiotic or a selectable marker. 2. From a mouse morula (a very young embryo consisting of a ball of undifferentiated cells), stem cells are isolated; these can be grown in vitro. For this example, we will take a stem cell from a white mouse. 3. The stem cells from step 2 are combined with the new sequence from step 1. This is done via electroporation (using electricity to transfer the DNA across the cell membrane). Some of the electroporated stem cells will incorporate the new sequence into their chromosomes in place of the old gene; this is called homologous recombination. The reason for this process is that the new and the old sequence are very similar. Using the antibiotic from step 1, those stem cells that actually did incorporate the new sequence can be quickly isolated from those that did not. 4. The stem cells from step 3 are inserted into mouse blastocyst cells. For this example, we use blastocysts from a grey mouse. These blastocysts are then implanted into the uterus of female mice, to complete the pregnancy. The blastocysts contain two types of stem cells: the original ones (grey mouse), and the newly engineered ones (white mouse). The newborn mice will therefore be chimeras: parts of their bodies result from the original stem cells, other parts result from the engineered stem cells. Their furs will show patches of white and grey. 5. Newborn mice are only useful if the newly engineered sequence was incorporated into the germ cells (egg or sperm cells). So we cross these new mice with others and watch for offspring that are all white. These are then further inbred to produce mice that carry no functional copy of the original gene. Limitations Create PDF with GO2PDF for free, if you wish to remove this line, click here to buy Virtual PDF Printer While knockout mice technology represents a valuable research tool, some important limitations exist. About 15 percent of gene knockouts are developmentally lethal, which means that the genetically altered embryos cannot grow into adult mice. The lack of adult mice limits studies to embryonic development and often makes it more difficult to determine a gene's function in relation to human health. In some instances, the gene may serve a different function in adults than in developing embryos. Knocking out a gene also may fail to produce an observable change in a mouse or may even produce different characteristics from those observed in humans in which the same gene is inactivated. For example, mutations in the p53 gene are associated with more than half of human cancers and often lead to tumours in a particular set of tissues. However, when the p53 gene is knocked out in mice, the animals develop tumours in a different array of tissues. There is variability in the whole procedure depending largely on the strain from which the stem cells have been derived. Generally cells derived from strain 129 are used. This specific strain is not suitable for many experiments (e.g., behavioural), so it is very common to backcross the offspring to other strains. Some genomic loci have been proven very difficult to knock out. Reasons might be the presence of repetitive sequences, extensive DNA methylation, or heterochromatin. Create PDF with GO2PDF for free, if you wish to remove this line, click here to buy Virtual PDF Printer