* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download complex I
Vectors in gene therapy wikipedia , lookup
Biosynthesis wikipedia , lookup
Photosynthesis wikipedia , lookup
Lipid signaling wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Magnesium transporter wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Signal transduction wikipedia , lookup
Microbial metabolism wikipedia , lookup
SNARE (protein) wikipedia , lookup
Proteolysis wikipedia , lookup
Metalloprotein wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Western blot wikipedia , lookup
Biochemistry wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Mitochondrial replacement therapy wikipedia , lookup
Citric acid cycle wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Mitochondrion wikipedia , lookup
Electron transport chain wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
MITOCHONDRIA
Speaker: 陳玉怜
Mar. 2017
1. Structure
2. Genetic system
3. Function: ATP synthesis
4. Dysfunction: cell apoptosis
eukaryotic cells
mitochondrion
mitochondria
• the specialized membranes
inside energy-converting
organelles
• are employed for the production
of ATP.
* occupy a substantial portion of the
cytoplasmic volume
* the metabolism of sugars is
completed: the pyruvate is
imported into the mitochondrion and
oxidized by O2 to CO2 and H2O.
(This allows 15 times more ATP to
be made than that produced by
glycolysis alone).
* stiff, elongated cylinders (a diameter of 0.5-1 µm)
* are remarkably mobile and plastic organelles
* move - often seem to be associated with
microtubules.
* remain fixed in one position where they
provide ATP directly to a site of high ATP
consumption
(packed between adjacent myofibrils in a
cardiac muscle cell; wrapped rightly around
the flagellum in a sperm)
Mitochondrial
plasticity
-time-lapse
microcinematography
-rapid changes of
shape in a living
cell
Dynamic mitochondrial reticulum
* form a continuous reticulum underlying the plasma
membrane
* a balance of fission and fusion determines the
arrangement of the mitochondria
* shape change; the network is constantly
remodeled by fission and fusion.
Mitochondrial
fission and fusion
-Involve both outer
and inner
mitochondrial
membrane
(movie)
The relationship between
mitochondria and
microtubules
Mitochondria ⇒
microtubules ⇒
The mitochondria tend to
be aligned along microtubules.
Localization of mitochondria near sites of
high ATP utilization in cardiac muscle
Localization of mitochondria near
sites of high ATP utilization in a
sperm tail
The mitochondrion:
*an outer membrane
*an inner membrane
*two internal
compartments (matrix and
intermembrane space)
Biochemical fractionation of
purified mitochondria into
separate components
The outer membrane
a transport protein
(porin) -forms large
aqueous channels
through the lipid bilayer.
-
- a sieve: all molecules of
5000 daltons or less (small
proteins). Such molecules
can enter the
intermembrane space, but
most of them cannot pass
the impermeable inner
membrane.
the inner membrane
Its lipid bilayer contains a
high proportion of the
"double" phospholipid
cardiolipin (has four
fatty acids rather than
two and may help to
make the membrane
especially impermeable
to ions)
Inner membrane
- contains a variety of transport proteins
that make it selectively permeable to
those small molecules that are
metabolized or required by the many
mitochondrial enzymes concentrated in
the matrix.
- enzymes of the respiratory chain, are
essential to the process of oxidative
phosphorylation, which generates most
of the animal cell's ATP.
cristae
(movie)
* The inner membrane is usually highly
convoluted, forming a series of
infoldings that project into the matrix.
•increase the area of the inner
membrane
* a liver cell: constitutes about onethird of the total cell membrane.
* cardiac muscle cells: many cristae
Matrix
The matrix enzymes include
those that metabolize
pyruvate and fatty acids to
produce acetyl CoA and
those that oxidize acetyl CoA
in the citric acid cycle.
Matrix granules
* Phospholipid
* Glycoprotein
* Lipid
* Calcium precipitable
lipoprotein
* Cytochrome c
oxidase
1. Structure
2. Genetic system
3. Function: ATP synthesis
4. Dysfunction: cell apoptosis
THE GENETIC SYSTEMS OF MITOCHONDRIA
-contain their own genomes, as well as their own biosynthetic machinery
for making RNA and organelle proteins.
Red: nuclear genome
Bright yellow spots:
mitochondrial genome
Green: mitochondrial
matrix space
(A) immunogold labelling with anti-DNA;
gold particles marking mtDNA are found
near the mitochondrial membrane. (B)
Whole mount view of and immunogold
labelling with anti-DNA; mtDNA (marked
by gold particles) resists extraction.
Mitochondria
contain
complete
genetic
systems
X
the process of
DNA replication
Why do
mitochondria
have their
own genetic
systems?
Two separate genetic systems — one in the
organelle and one in the cell nucleus.
Most of the proteins - by nuclear DNA, synthesized
in the cytosol, and then imported individually into
the organelle.
Some organelle proteins and RNAs - the organelle
DNA and are synthesized in the organelle itself.
The human mitochondrial genome
contains about 16500 nucleotides and
encodes 2 ribosomal RNAs, 22 transfer RNAs,
and 13 different polypeptide chains.
1. Structure
2. Genetic system
3. Function:
ATP synthesis
4. Dysfunction: cell apoptosis
Chemiosmotic coupling (chemiosmosis)
* the chemical bond-forming reactions
that generate ATP ("chemi")
* membrane-transport processes
("osmotic").
* The coupling process occurs in two linked
stages, both of which are performed by
protein complexes embedded in a
membrane.
proton-motive force
High-energy electrons are generated
via the citric acid cycle
Fuel: pyruvate (from glucose and other
sugars) and fatty acids (from fats)
-are transported across the inner
mitochondrial membrane and then
converted to the crucial metabolic
intermediate acetyl CoA by enzymes
located in the mitochondrial matrix.
The acetyl groups in acetyl CoA are then
oxidized in the matrix via the citric acid
cycle.
Energy –generating
metabolism
in mitochondria
NADH: reduced
nicotinamide adenine
dinucleotide
Pyruvate+HS-CoA+NAD+
→acetyl CoA+CO2+NADH+H+
decarboxylation
The tricarboxylic acid cycle (TCA cycle)
Nicotinamide
adenine
dinucleotide
2C
4C
6C
4C
6C
4C
4C
5C
4C
flavin adenine
dinucleotide reduced
Acetyl CoA+2H2O+FAD+3NAD++GDP+Pi→ 2CO2+FADH2+3NADH+3H++GTP+HS-CoA
Spectroscopic Methods Have Been Used to Identify
Many Electron Carriers in the Respiratory Chain
@ cytochromes a, b, and
c.
- constitute a family of
colored proteins that are
related by the presence of a
bound heme group, whose
iron atom changes from the
ferric oxidation state (Fe 3+ )
to the ferrous oxidation
state (Fe 2+ ) whenever it
accepts an electron.
heme group = a porphyrin
ring +iron atom +four
nitrogen atoms
@ Iron-sulfur proteins
either two or four iron atoms are bound to an equal
number of sulfur atoms and to cysteine side chains,
forming an iron-sulfur center on the protein.
@ ubiquinone
The simplest of the electron carriers in the respiratory chain—
and the only one that is not part of a protein—is a small
hydrophobic molecule that is freely mobile in the lipid bilayer
A quinone (Q) can pick up or donate either one or two electrons;
upon reduction, it picks up a proton from the medium along with
each electron it carries.
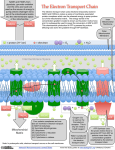
The Respiratory Chain Includes Three Large Enzyme
Complexes Embedded in the Inner Membrane
1. The NADH dehydrogenase complex (complex I) is
the largest of the respiratory enzyme complexes, containing more
than 40 polypeptide chains. It accepts electrons from NADH and
passes them through a flavin and at least seven iron-sulfur centers to
ubiquinone. Ubiquinone then transfers its electrons to a second
respiratory enzyme complex, the cytochrome b-c1 complex.
2. The cytochrome b-c1 complex contains at least 11 different
polypeptide chains and functions as a dimer. Each monomer contains
three hemes bound to cytochromes and an iron-sulfur protein. The
complex accepts electrons from ubiquinone and passes them on to
cytochrome c, which carries its electron to the cytochrome oxidase
complex.
3. The cytochrome oxidase complex also functions as a dimer;
each monomer contains 13 different polypeptide chains, including two
cytochromes and two copper atoms. The complex accepts one electron at a
time from cytochrome c and passes them four at a time to oxygen.
* During the transfer of electrons from NADH to oxygen,
ubiquinone and cytochrome c serve as mobile carriers.
* Protons are pumped across the membrane by each of the
respiratory enzyme complexes.
How electrons are donated by NADH
A hydride ion (H- a hydrogen atom and
extra electron)
NADH: reduced nicotinamide adenine dinucleotide
NAD+: nicotinamide adenine dinucleotide
The process of electron transport
- The hydride ion is removed from NADH
(to regenerate NAD+) and is converted
into a proton and two electrons (H- ⇒ H+
+ 2e-).
- Each of these ions being tightly bound
to a protein molecule that alters the
electron affinity of the metal ion. Most of
the proteins involved are grouped into
three large respiratory enzyme
complexes.
A chemiosmotic process
converts oxidation energy into ATP
These electrons, carried by NADH and
FADH2, are then combined with O2 by
means of the respiratory chain
embedded in the inner mitochondrial
membrane. The large amount of
energy released is harnessed by the
inner membrane to drive the conversion
of ADP + Pi to ATP (oxidative
phosphorylation).
The NADH dehydrogenase complex (complex I)
-It accepts electrons from NADH and passes them through a flavin and at least seven
iron-sulfur centers to ubiquinone.
- Ubiquinone then transfers its electrons to a second respiratory enzyme complex, the
cytochrome b-c1 complex.
The cytochrome b-c1 complex
- Each monomer contains three hemes bound to cytochromes and an ironsulfur protein.
- The complex accepts electrons from ubiquinone and passes them on to
cytochrome c, which carries its electron to the cytochrome oxidase complex.
The cytochrome b-c1 complex
contains at least 11 different polypeptide chains and functions as a dimer.
Greencytochrome b
BlueCytochrome c1
PurpleIron-sulfur center
X-ray
The atomic structure of cytochrome b-c1
The cytochrome oxidase complex
-including two cytochromes and two copper atoms.
-The complex accepts one electron at a time from cytochrome c
and passes them four at a time to oxygen.
The molecular
structure of
cytochrome oxidase
- a dimer
- each monomer
contains 13 different
polypeptide chains,
including two
cytochromes and two
copper atoms.
An iron-copper center in cytochrome oxidase catalyzes O2 reduction
A general model for H+ pumping
Redox potential changes along the mitochondrial electron-transport chain
electron
transport chain
= the entire set
of proteins in
mem+ small
molecules in
electron transfer
Harnessing energy for life
Chemiosmotic coupling
*Electrochemical proton
gradient-drive other memembedded protein
machines
*Special proteins couple
the “downhill” H+ flow to the
transport of specific
metabolites into and out of
the organelles
*Electrochemical proton
gradientrapid rotation of the
bacterial flagellum
As electrons move along the respiratory chain, energy stored
as an electrochemical proton gradient across the inner
membrane
- a voltage gradient (the inside negative and the outside
positive)
- a pH gradient (the pH higher in the matrix than in the
cytosol
the ∆pH and
the ∆Vconstitute
electrochemi
cal proton
gradient
How the Proton Gradient Drives ATP Synthesis
The electrochemical proton gradient across the
inner mitochondrial membrane is used to drive
ATP synthesis in the critical process of oxidative
phosphorylation.
The major net energy conversion
catalyzed by the mitochondrion
ATP synthase
head portion -F1
ATPase
(matrix)
F0- transmembrane
H+ carrier
-inner
mitochondrial
membrane
ATP synthase (F0F1ATPase, more than 500,000 daltons)
-a lollipop head and composed of a ring of 6 subunits, projects on the
matrix side of the inner mitochondrial membrane.
a "stator“ (green)
a "rotor" -is formed by a ring of 10 to 14 identical transmembrane protein
subunits. (Paul Boyer &John E. Walker: 1997 Nobel Prize in Chemistry)
How the Proton Gradient Drives Coupled
Transport Across the Inner Membrane
In mitochondria, many charged small molecules,
such as pyruvate, ADP and Pi, are pumped into
the matrix from the cytosol, while others, such as
ATP must be moved in the opposite direction.
•Pyruvate and inorganic phosphate (Pi) are cotransported inward with H+ as the H+ moves into
the matrix.
•ADP-ATP co-transport is driven by the voltage
difference across the membrane.
Some of the active transport processes driven by the electrochemical
proton gradient across the inner mitochondrial membrane
Proton Gradients Produce Most of the
Cell's ATP
The vast majority of the ATP produced from
the oxidation of glucose in an animal cell is
produced by chemiosmotic mechanisms in
the mitochondrial membrane. Oxidative
phosphorylation in the mitochondrion also
produces a large amount of ATP from the
NADH and the FADH2 that is derived from
the oxidation of fats.
ATP Synthase Can Also Function in Reverse to Hydrolyze ATP and Pump H+
In addition to harnessing the How of H+ down an electrochemical proton gradient to
make ATP, the ATP synthase can work in reverse: it can use the energy of ATP
hydrolysis to pump H+ across the inner mitochondrial membrane. It thus acts as a
reversible coupling device, interconverting electrochemical proton gradient and
chemical bond energies.
As protons pass through a narrow channel
formed at the stator-rotor contact, their
movement causes the rotor ring to spin. This
spinning also turns a stalk attached to the
rotor.
1. Structure
2. Genetic system
3. Function: ATP synthesis
4. Dysfunction:
cell apoptosis
Mitochondrial dysfunction
Cause cell death through ATP
depletion and Ca 2+ dysregulation.
Proteins released from mitochondria
(cytochrome c, apoptosis-inducing
factor, smac-diablo)
Be modulated by regulated targeting
of BCL2 family members to the outer
mitochondrial membrane.
BCL2 family
* Antiapoptotic members (Bcl-2, Bcl-xL, Bclw, Mcl-1, A1)
* BH1-BH4
* Proapoptotic members (Bax, Bak, Bok,
Bcl-rambo, Bcl-GBID)
BH1-BH3
* BH3-only protein (numerous pro-apoptotic
members)(Bad, Bid, Bim, Blk)
Schematic drawing of Bcl-2
and its related proteins
Activation of Bax/Bak by BH3-only proteins.
Once activated, BH-3 only proteins are translocated
to the mitochondria to inactivate anti-apototic members
of the Bcl-2 family and activate multi-domain members,
resulting in an increase of outer mitochondrial
membrane permeability.
Bad, Bid, Bim, Blk
Mechanisms for MOMP during apoptosis
ANT: adenine nucleotide transporter
VDAC: voltage-dependent anion channel
PT: permeability transition
Green and Kroemer, Science, 2004
Apoptotic signal transduction pathways
Apoptotic protease-activating factor 1
Tsujimoto, Journal of cellular physiology, 2003
Release of mitochondrial proteins by formation of apoptotic proteins-conducting pores
X
X
X
X
X
Moll et al., 2006
The mitochondrial pro-apoptotic activities of p53
EXR: retinoid X receptor
The Nur77-Bcl2 apoptotic and survival pathways
DBD:DNA binding domain
Mechanistic similarities of the transcription-independent
pro-death program of p53 and Nut 77 at mitochondria
Examples of pathogenic processes involving excessive or deficient MOMP
Disease
Pathogenic perturbation of
MOMP
Pharmacological correction of
deregulated MOMP
Ischemia
reperfusion
damage of brain
or heart
Redox stress, excessive Ca2+
load, absent adenosine
triphosphate and nicotine
adenine dinucleotide, and
accumulating fatty acids favor
PT and MOMP.
Bcl-2 inhibitors of the PT pore,
as well as mito KATP channel
openers, can exert neuro- or
cardioprotective effects.
Neurodegenerat
ive diseases
Respiratory dysfunction
Putative inhibitors of the PT
affects highly sensitive
pore (minocyclin, rasagiline,
neurons in the central nervous and tauroursodeoxycholic acid)
system, leading to their
can prevent
premature death.
neurodegeneration.
Liver disease
Hepatotoxins (including bile
Ursodeoxycholic acid prevents
acid and ethanol) and hepatitis bile acid—induced PT and thus
B or C—encoded proteins
exerts hepatoprotective
induce MOMP.
effects.
Cancer
MOMP-inhibitory proteins from
the Bcl-2 family or unrelated
proteins (such as Muc1) enhance
apoptosis resistance.
Cytotoxic agents targeting Bcl2—like proteins, PT pore
components, and/or
mitochondrial lipids enforce
MOMP and kill cancer cells.
1. Structure
2. Genetic system
3. Function: ATP synthesis
4. Dysfunction: cell apoptosis