* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Non-small-cell lung carcinoma
Pathogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Point mutation wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenomics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Minimal genome wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Human genome wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Public health genomics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genomic library wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Copy-number variation wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Helitron (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome editing wikipedia , lookup
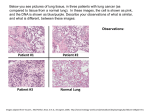
BIF Non-small-cell lung carcinoma Non-small-cell lung carcinoma (NSCLC) is any type of epithelial lung cancer other than small cell lung carcinoma (SCLC). As a class, NSCLCs are relatively insensitive to chemotherapy, compared to small cell carcinoma. The most common types of NSCLC are squamous cell carcinoma(鳞状细胞癌), large cell carcinoma(大细胞癌), and adenocarcinoma(腺癌) but there are several other types that occur less frequently, and all types can occur in unusual histologic variants and as mixed celltype combinations. 2013 BIF Lung adenocarcinoma Adenocarcinoma of the lung is currently the most common type of lung cancer in "never smokers". Adenocarcinomas account for approximately 40% of lung cancers. Squamous cell lung carcinoma Squamous cell carcinoma (SCC) of the lung is more common in men than in women. It is closely correlated with a history of tobacco smoking, more so than most other types of lung cancer. Large-cell lung carcinoma Large cell lung carcinoma (LCLC) is a heterogeneous group of undifferentiated malignant neoplasms originating from transformed epithelial cells in the lung 2013 BIF Copy-number variation Copy-number variations (CNVs)—a form of structural variation—are alterations of the DNA of a genome that results in the cell having an abnormal number of copies of one or more sections of the DNA. CNVs contrast with singlenucleotide polymorphisms (SNPs), which affect only one single nucleotide base. 2013 BIF Identification Copy number variation can be discovered by cytogenetic techniques 1. fluorescent in situ hybridization(荧光原位杂交) 2. comparative genomic hybridization 3. array comparative genomic hybridization 4. virtual karyotyping with SNP arrays. 2013 BIF 1. fluorescent in situ hybridization FISH样本的制备 →探针的制备 →探针标记 →杂交 →(染色体显带) →荧光显微镜检测 →结果分析。 2013 2. comparative genomic hybridization CGH 只能检测不平衡的 染色体改变 2013 BIF BIF 3. Array comparative genome hybridization (aCGH) 1. DNA from a test sample and normal reference sample are labelled differentially, using different fluorophores, 2. hybridized to several thousand probes. The probes are derived from most of the known genes and non-coding regions of the genome, printed on a glass slide. 3. The fluorescence intensity of the test and of the reference DNA is then measured, to calculate the ratio between them and subsequently the copy number changes for a particular location in the genome. 2013 BIF Prevalence in humans DNA copy number variation is a widespread and common phenomenon among humans Role in disease some CNVs have been associated with susceptibility or resistance to disease Gene copy number can be elevated in cancer cells. the EGFR copy number can be higher than normal in non-small cell lung cancer Datasetes 2013 BIF Copy-number variation: the end of the human genome? Paul H. Dear Trends in Biotechnology 2009 2013 文献内容: BIF 1. 简略提Copy-number variation的概念,鉴定的 方法,及简要的历史 2. Copy-number variation和phenotype 3. Copy-number changes 和 cancer 4. MCC and mMCC – a digital genomics approach to CNVs 5. Location, location, location! 6. Somatic CNV 2013 BIF CNVs and phenotype Almost by definition, most CNVs have either no phenotypic consequences or only subtle or benign ones they are comparable to the majority of SNPs some CNVs do indeed have phenotypic consequences CNVs in genes known to be implicated in disease 2013 BIF Copy-number changes and cancer CNVs have clearly been shown to have the potential to indirectly influence a healthy individual’s susceptibility to cancer,for example by varying the gene dosage of tumour suppressors or oncogenes Changes in copy number of parts of the genome are known to be a feature of many cancers it difficult to determine which copy-number changes are significant or causative and which are purely incidental or of minor importance. 2013 MCC and uMCC – a digital genomics approach to CNVs molecular copy-number counting 2013 BIF BIF The initial results, however, were distorted owing to the extreme fragmentation of the DNA; as with any PCRbased method, longer target sequences are more susceptible than shorter ones to destruction caused by DNA damage by choosing targets of short and approximately uniform length, this problem could be overcome, and this led to the modified method of uMCC. 2013 BIF Location, location, location! In a few cases, usually those showing large structural abnormalities, the whereabouts of the extra copies are known, but in most cases they are not. Indeed, it is extremely difficult to determine where the further copies of a CNV are located in the genome unless they are large enough to be discerned by fluorescence in situ hybridization. 2013 BIF Somatic CNV Copy-number differences between somatic cells could potentially be quite widespread without our being aware of it. The authors used CGH to reveal copy-number differences between different tissues that were sample dpost-mortem from single individuals who had died from apparently non-genomic causes. 2013 BIF Integrated analysis of DNA copy number and gene expression microarray data using gene sets Renée X Menezes BMC Bioinformatics 2009 2013 BIF propose to look for associations between copy number and expression not only using individual genes, but also using gene sets. We wish to find which individual copy number changes affect gene expression levels within the same chromosomal region. 2013 BIF Methods The gene-to-gene model consider copy number as the dependent variable, so expression is handled as the independent variable. The simplest model to consider is 2013 BIF 2013 BIF The gene-set model An ideal way to avoid these problems is to include in the model all expression probes within the same large region. This leads us to the model, for each copy number probe i, 2013