* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Polypeptides and Proteins
Nucleic acid analogue wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Magnesium transporter wikipedia , lookup
Butyric acid wikipedia , lookup
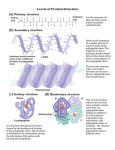
Gene expression wikipedia , lookup
Western blot wikipedia , lookup
Protein moonlighting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Circular dichroism wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Protein domain wikipedia , lookup
Protein adsorption wikipedia , lookup
List of types of proteins wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Peptide synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
Bottromycin wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
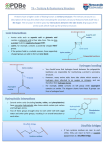
Polypeptides and Proteins These molecules are composed, at least in part, of chains of amino acids. Each amino acid is joined to the next one through an amide or peptide bond from the carbonyl carbon of one amino acid “residue” to the α-amino group of the next. At one end of the chain there will be a free or protonated amino group: the N-terminus. At the other end there will be a free carboxyl or carboxylate group: the C-terminus. By convention the N-terminus is drawn at the left end, as shown below in a generic hexapeptide (6 amino acid residues). H H3N C C R O N-terminus H H H H H H H H H H N C C N C C N C C N C C N C R O R O R O R O amino acid residue planar sections C O R O C-terminus The peptide linkage is planar --H C C O H N C C C N O C Note that the backbone chain of amino acid residues consists of a series of flat “plates” that “join” at the α-carbons of the amino acid residues. It is the tetrahedral α-carbons that allow the chain to bend and have some flexibility. The plates are flat because the amide (or 1 peptide) C-N bond is planar owing to its double bond character as shown by resonance in the figure above. Because peptide molecules are large a shorthand method is used to indicate their structures, eg, bradykinin — H H H3N C C O N C H C N C O H H C N C O H H C N C H O C CH2 O NH C HN NH2 H H N C H C N C CH2 O H H C O N C H H C N C C O CH2 O O OH NH C HN NH2 As you can see, it is simpler to use the three letter abbreviations for the amino acid residues than it is to draw the usual structure: Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg. 2 Note that the N-terminus is at the left end and the C-terminus is at the right end; it is critical that we hew to this convention when writing structures using the three letter abbreviations — Arg-Phe-Pro-Ser-Phe-Gly-Pro-Pro-Arg would be a different molecule. Determination of a polypeptide structure — 1. Which amino acid residues are present? 2. How many of each? 3. What is their sequence? (1) may be determined by complete acid hydrolysis and qualitative analysis of the hydrolysate. (2) may be determined by complete acid hydrolysis, quantitative analysis of the hydrolysate, and the determination of the MW of the polypeptide. Consider a polypeptide, MW = 637. Complete hydrolysis of 637 mg (1.00 mmole) gives: Asp, 133 mg (1.00 mmole); Phe, 165 mg (1.00 mmole); Val, 117 mg (1.00 mmole); Glu, 294 mg (2.00 mmole). Therefore, polypeptide = AspGlu2-Phe-Val, not in sequence. The sequence (3) may be determined by partial hydrolysis. Suppose the following units (among others) are found after partial hydrolysis of the above polypeptide: Asp-Glu, Val-Asp, Phe-Val, Glu-Glu. We can extrapolate to the overall sequence as follows: 3 Asp-Glu Val-Asp Phe-Val Glu-Glu Phe-Val-Asp-Glu-Glu Therefore, the sequence is Phe-Val-Asp-Glu-Glu. Another --- real life --- example would be the determination of the sequence of amino acid residues in the B-chain of insulin. [Insulin consists of two polypeptide chains, A and B, held together by two disulfide bridges, C-S-S-C. A disulfide bridge results from oxidative coupling of the S-H groups of two cysteine residues.] S S S S First the disulfide bridges would be cleaved A reductively, regenerating cysteine SH groups, B and separating the two chains. Then the chain of interest, B in this case, would be partially hydrolyzed. The result obtained when this was done follows: Phe-Val-Asn-Gln-His-Leu-Cys-Gly-Ser-His-Leu Ser-His-Leu-Val Leu-Val-Glu-Ala Val-Glu-Ala-Leu Ala-Leu-Tyr Tyr-Leu-Val-Cys Val-Cys-Gly-Glu-Arg-Gly-Phe Gly-Phe-Phe-Tyr-Thr-Pro-Lys Tyr-Thr-Pro-Lys-Ala _______________________________________________________________________________________________________________________ Phe-Val-Asn-Gln-His-Leu-Cys-Gly-Ser-His-Leu-Val-Glu-Ala-Leu-Tyr-Leu-Val-Cys-Gly-Glu-Arg-Gly-Phe-Phe-Tyr-Thr-Pro-Lys-Ala 4 But how is the sequence in the fragments determined? Answer: Terminal residue analysis. Some reagents will react specifically with one terminus or the other, thus enabling one to determine the nature of the terminal residue. In some cases, the terminal residue can be cleaved off and the remainder of the polypeptide remains (more or less) intact. The remaining polypeptide can be subjected to terminal residue analysis, etc. The Edman Degradation — base H2NCHCNHCHC + Ph N C S phenyl isothiocyanate R O R' polypeptide H H H2O Ph N C NCHC NHCHC HCl R O R' O S N-terminal tagged polypeptide S H C Ph N N H NHCHC + C C H R' O O R degraded peptide One of 20 possible phenylthiohydantoins, identified via chromatography, thereby identifying the terminal a a residue. 5 Polypeptides : Smaller than proteins. MW < ~7-10,000 or < ~50 amino acid residues. Polypeptides are usually flexible: they usually do not have the secondary structure characteristic of proteins. (Nor do they have tertiary or quarternary structure.) Examples: bradykinin: helps regulate blood pressure; pain. methionine enkephalin: Tyr-Gly-Gly-Phe-Met, morphine like analgesic. oxytocin: S S cys-tyr-ileu-gln-asn-cys-pro-leu-gly-NH2 causes uterine contraction and lactation; not species specific— labor induced via chicken oxytocin. vasopressin (note the structural similarity to oxytocin): S S cys-tyr-phe-gln-asn-cys-pro-arg-gly-NH2 6 antidiuretic. Proteins — Classification by Gross Chemical Structure Simple: consist only of amino acid residues. Conjugated: consist of polypeptide portion(s) and nonpolypeptide portion(s) called prosthetic group(s), eg lipoproteins, glycoproteins, nucleoproteins, metalloproteins. Classification by Gross Morphology Fibrous: insoluble in water, resist hydrolysis, eg, sericine (silk fibroin), hard keratin (hair, nails, claws), soft keratin (skin), myosin (muscle), collagen (connective tissue, bones). Globular: soluble or colloidal in water, eg, hemoglobin, enzymes, antibodies, some hormones. 7 Protein Structure Primary: sequence of amino acid residues. Secondary: way in which segments of polypeptide backbone are oriented in space; depends on planarity of peptide bond and, often, Hbonding. The only places in the protein chain where there is torsional flexibility is at the α-carbons: the angles Φ and Ψ can change causing one plane to rotate relative to the other. One could imagine that all of the planar “plates” in the protein backbone lie in the same plane. This would produce a flat strand. By snaking back and forth other parts of the strand could lie down alongside the first in parallel (or antiparallel) fashion creating a flat sheet. Or other protein molecules could lie down parallel (or antiparallel) to the first. 8 This flat sheet would allow hydrogen bonding between adjacent strands of the protein (favorable for formation of the sheet). However, the R groups sterically interfere with each other in this setup. This hypothetical structure has not been observed in nature. Antiparallel β-pleated sheet. Black = C, white = H, blue = N, Red = O, Yellow = R (side chain). The steric interference between the R groups in the flat sheet can be reduced if the sheet becomes “pleated”. In this arrangement the angle between adjacent plates is not zero. Hydrogen bonding is still possible and now the R groups can splay away from each other to some extent. This setup works if the R groups are small. This is the case for silk fibroin which does form the pleated sheet structure. 9 Another geometry which is possible is the helix. This arrangement allows hydrogen bonding in directions roughly parallel to the helical axis (dotted bonds) and the R groups are oriented outward from the helix, away from each other. This happens in nature. Several varieties of helix are known; the most common form is known as the α-helix. It is a right-handed helix (if it were a wood screw, turning it clockwise would cause it to be driven into the wood). [The mirror image of a right-handed helix is a left handed one. The mirror images of the L-amino acids in the right-handed helix would be D-amino acids. The chirality of the amino acid residues determines the chirality of the helix!] α-Helix. Black = C, white = H, blue = N, red = O, purple = R. 10 Tertiary: way in which protein as a whole is twisted in three dimensions; may depend on disulfide bonds, Hbonds, electrostatic attractions, hydrophobic interactions. The principal factors affecting protein tertiary structure. Quaternary: aggregate structures held together by nonbonded interactions or occasionally disulfide links. Let's look at some proteins! 11 The Primary Structure of Bovine Insulin — Insulin is a protein hormone produced in the pancreas. It is essential for the regulation of carbohydrate metabolism. Amino Acid Residues Species A-8 A-9 A-10 B-30 Cow Ala Ser Val Ala Sheep Ala Gly Val Ala Horse Thr Gly Ileu Ala Human Thr Ser Ileu Thr Pig Thr Ser Ileu Ala Whale Thr Ser Ileu Ala 12 I have never seen a discussion of the secondary or tertiary structure of insulin. It is a small protein so its secondary structure may be somewhat flexible, although one would expect it to have conformations of lower and higher energy with one or several of lower energy predominating. With regard to tertiary structure the disulfide links would certainly be important. Collagen — Collagen is found in all multicellular animals and is the most abundant protein of vertebrates. It is a fibrous protein, the fibers of which have great tensile strength. It is found in the connective tissue of teeth, bones, ligaments, tendons and cartilage. Primary structure — Collagen is rich in glycine (R=H). This enables the polypeptide strands to fit tightly to other strands without steric difficulties. The other major components are proline and hydroxyproline. Secondary structure — A left-handed helix. Tertiary structure — A right-handed helix. 13 Quaternary structure — A cabling of three right-handed helicies. However, this continues to higher levels of organization as we can see in the following photomicrographs. 14 Hemoglobin — Hemoglobin is a complex, conjugated, metalloprotein. It is what makes red blood cells red. It carries oxygen throughout the body from the lungs. Each hemoglobin protein consists of four units, two α-units and two β-units. This is its quaternary structure. Each unit consists of a globin and a heme. The globin is a chain of amino acid O Oxygen residues. The heme is O H C CH CH a porphyrin containing CH one iron atom. The iron CH HC N N ++ atom carries the oxygen Fe Heme N N molecule. The CH HC presence of the nonCH CH COO OOCCH CH N polypeptide heme makes hemoglobin a N H CH conjugated protein. Histidine A A O CH Residue Each type of unit, α and HN C of Globin β, has its own particular shape composed of various twists and turns. This is its tertiary structure. 2 3 2 3 3 3 - - 2 2 2 2 2 Within each unit the polypeptide backbone adopts certain conformations going down the chain – in many places this is a helix. This is the secondary structure of the protein. 15 Finally, the sequence of the amino acids in the α- (141 residues) and β-globins (146 residues) is the primary structure. Oxygenated hemoglobin. The amino acid residues are shown by number in each globin, starting with #1 at the N-terminus. This image was drawn looking down the C-2 axis. Ignore the large grey arrows. 16 Over 100 abnormal varieties of hemoglobin have been reported in humans. The abnormalities are in the globins. One of these is the replacement of the glutamic acid residue at position #6 in the β-globins by a valine residue: Normal: Val-His-Leu-Thr-Pro-Glu-Lys ---> 146 Abnormal: Val-His-Leu-Thr-Pro-Val-Lys ---> 146 This change causes the hemes associated with the βglobins to be unable to carry oxygen. As a result cells become oxygen starved. This alteration also causes the hemoglobin to crystallize and this, in turn, causes some red blood cells to stiffen and deform into a crescent shape. These cells can block blood flow through capillaries. Furthermore, the body recognizes these cells as abnormal and destroys them, resulting in anemia. (Life of abnormal cells ~ 16 days; normal ~ 120 days.) This condition, along with the shape of the cells gives rise to the name of this genetic disease: sickle cell anemia. 17 People who have inherited two sickle cell genes (one from mother, one from father) will have the disease. Those who have inherited one normal and one defective gene will have some abnormal hemoglobin, but usually live normal lives; they are said to have the trait, and can pass the gene on to their children. 18 q s N,s N,s y y y y N,N N,s N,s s,s 19 N=normal gene s=defective gene N,N=completely normal N,s=sickle trait s,s=sickle cell disease Why hasn't this gene disappeared from the gene pool? Wouldn't it be removed by (Darwinian) natural selection? Who gets the s gene? People who live, or whose ancestors lived, in areas where malaria is, or was, prevalent. Malaria? The malarial parasite spends part of its life cycle in red blood cells. People who are N,s are somewhat resistant to malaria. Thus, the presence of malaria confers some advantage to the s gene. This gene did not survive longterm in populations where malaria is not endemic. The map below shows the relationship between malaria and the presence of the sickle gene. 20 Map of Misery 21