* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mass spectrometry and stable isotope labeling for
Nutriepigenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Hammerhead ribozyme wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetically modified organism containment and escape wikipedia , lookup
Genetic engineering wikipedia , lookup
Transfer RNA wikipedia , lookup
Genetically modified food wikipedia , lookup
Metagenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genetic code wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Messenger RNA wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
RNA interference wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Polyadenylation wikipedia , lookup
Primary transcript wikipedia , lookup
RNA-binding protein wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
RNA silencing wikipedia , lookup
History of RNA biology wikipedia , lookup
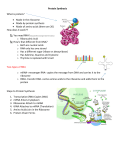
Mass spectrometry and stable isotope labeling for quantitative analysis of ribosomal RNA modifications. Anna M. Popova, James R. Williamson The Scripps Research Institute, Department of Integrative Structural and Computational Biology, 10550 N. Torrey Pines Rd., La Jolla, CA, 92037. Posttranscriptional modifications provide a unique and often unappreciated way to induce control over RNA structure, metabolism and biological functions. Over the last years, the list of modified RNAs has expanded and increasing numbers of modified sites continue to be found in catalytic, non-coding and messenger RNAs across the species. Still, their biological implications remain elusive, in part, because of the lack of experimental methods that would enable quantitative assessment of modified RNA. Ribosomal RNA (rRNA) is a core component of the ribosome RNA-protein complex that translates genetic information into proteins. rRNA is extensively modified during the ribosome biogenesis process, with 30-200 modifications being introduced at functionally important domains of the ribosome, from bacteria to humans. To understand interplay between modifications and ribosome assembly we have developed quantitative Mass Spectrometry (qMS) technology to perform global analysis of modifications in abundant RNAs. Based on metabolic labeling of RNA with stable isotopes, chromatographic separation of rRNA nucleolytic fragments, and accurate fitting of the resulting isotope distributions (see Figure), qMS enabled us to profile 80% of all rRNA methylated sites and pseudouridines present in E. coli and S. Cerevisiae. Great sensitivity and automation of the method allowed us to simultaneously monitor many rRNA modifications even in low-abundant ribosome intermediates samples, permitting quantitative analysis of modifications with precision and accuracy not previously achieved using any available biochemical technique. I have been successfully using qMS technology to characterize temporal and functional dependencies between specific RNA modification steps and the ribosome assembly process in bacteria. Furthermore, we are advancing the technology for analysis of eukaryotic ribosomes (yeast and human) where the biogenesis pathway is significantly more complex, and where defects in RNA modifications are linked to genetic disorders and cancer. Overall, our research sets a stage for in depth exploration of rRNA modification pathways and provides important technical contributions to the emerging and poorly explored field of RNA modification biology.