* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Mouse_lecture
Human genome wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Gene desert wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Pathogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Public health genomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Medical genetics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
Genome editing wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
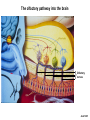
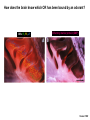
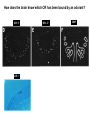
Vertebrate Genetics Nirao Shah Haploid facts of life Human Life Cycle Drive at 16 yrs Vote, marry, shoot at 18 yrs C2H5OH at 21 yrs ~5 days ~12 yrs ~7 days ~6 mths ~8 weeks ~39 weeks Sex determination in the mouse XX = female XY = male XO = female XXY = male What chromosome harbors the sex-determining locus? Y chromosome Single gene on Y, Sry, is the sex determining locus Mid-gestation embryo, E9.5 Bi-potential gonad Sry Testes Testosterone Bipotential body & brain Male phenotype Default Ovaries Estrogen & Progesterone Non-autonomous Female phenotype How know Sry is sex determining locus? Is it sufficient? Is it necessary? Forward genetics: Phenotype to genotype Reverse genetics: Genotype to phenotype Why do forward genetics? Why not forward genetics in vertebrates? The mouse: • is diploid • is not transparent • does not self-fertilize Screen for the phenotype you want, Unbiased, Saturation, Allelic series, Time ($), Space ($), Unlikely to saturate, Some problems not accessible (incl in flies and worms) How know Sry is sex determining locus? Is it sufficient? Is it necessary? Sufficiency - drive transgene in female and masculinize - rescue Sry null with transgene Necessity - knockout Sry (doesn’t work bc cannot recombine on Y) - deletions in the population Reverse genetics in the mouse Transgene – Random insertion of DNA construct in the genome reporters, toxins, RNAi, rescue, sufficiency, enhancer bash Targeted manipulation – homologous recombination mediated aka gene targeting, knock-in, knock-out Basic ingredients for a transgenic construct Generating a mouse bearing the desired transgene Fertilized eggs from donor WT females Inject your DNA Transfer injected eggs into WT recipient females Genotype progeny (why?, how?) Breed founder progeny harboring the transgene Check progeny of founders for expression of your transgene Why? How? Male pronuclear injection of transgene Typically integrates into genome as a multi-copy insertion No expression or mis-expression of your transgene: • Inadequate promoter, lacking regulatory elements • Position effect: local genomic context precludes correct expression Solutions: • Screen more progeny • BAC transgenic • Lentivirus transgene • Gene targeting BAC transgene • Bacterial artificial chromosome • ~150 kb genomic insert (publicly available library spanning the genome) • BAC libraries fully annotated, order by phone • Random integration • Lower freq of position effect • Gene regulatory information usually within the BAC you ordered • Usually single copy insertion What’s the construct? GENSAT collection of BAC transgenics – BAC mouse by phone (http://www.gensat.org/index.html) BAC transgene Chunks can break off during injection Incomplete regulatory information – misexpression (or more rarely, position effect) Lentiviral transgene • HIV backbone stripped of all viral genes (why?) • Recombinant and replication incompetent (why?) • Produced in tissue culture • Just soak fertilized eggs in virus and transfer to female • Position effect rare (Integration usually close/within active locus) • Pan-tropic (rats, voles.....) • Max insert size is 10 kb (why is this bad?) • Integration usually close/within active locus A genetic approach to understand how we recognize odors We recognize ~ 103 odors How does the nose detect so many odors? How does the brain know what the nose detects? The olfactory pathway into the brain } Olfactory nerves Axel 1995 Forward genetic screens in flies/worms failed to identify odorant receptors. Why? Redundancy in receptors Difficult behavioral screens Cloning olfactory receptors • Membrane preps of nose show • This requires GTP cAMP in presence of odorants In the late 1980s, Linda Buck & Richard Axel reasoned that, 1. odorant receptors must be GPCRs 2. these GPCRs must be encoded by a gene family 3. these GPCRs must be restricted to the nose Cloning olfactory receptors Buck & Axel designed degenerate PCR primers based on known GPCRs (PCR had just been invented!) Performed RT-PCR on mRNA from nose g t a c c a c atg gcc tat gat agg tat...... M A Y D R Y.... Cloning olfactory receptors * * GPCRs encoded by a gene family in the nose Cloning olfactory receptors GPCRs encoded by a gene family in the nose are not expressed elsewhere Cloning olfactory receptors ~1300 odorant GPCRs encoded in the genome ~5% of coding genes in genome Highly conserved across vertebrates Flies and worms have unrelated GPCR families of odorant receptors. Identified later from mining the sequenced genomes in mid-90s. Worm GPCRs identified by Emily Troemel in Bargmann lab at UCSF. How does the brain know which OR has been bound by an odorant? ~ 107 olfactory sensory neurons in mouse nose ~ 1300 ORs Could have combinatorial expression, several defined ORs per neuron Alternately, single OR gene per neuron. • In situ hybridization (ISH) reveals sparse expression of individual ORs (0.1 - 0.5% neurons) • ISH pattern additive (what does this mean?) • Although seemingly stochastic, symmetric expression across midline • Reproducible between individuals Ressler; Vassar 1993 How does the brain know which OR has been bound by an odorant? ORs: I7, F3, J7 Olfactory marker protein (OMP) 1 mm Vassar 1993 How does the brain know which OR has been bound by an odorant? OR: I7 OR: J7 OMP OR: I7 Vassar 1993 How does the brain know which OR has been bound by an odorant? Taken together, highly probable that each olf. sensory neuron expresses only one OR. Eventually confirmed by single cell RT-PCR. Serial dilution of dissociated MOE population reveals something astonishing: An olf. sensory neuron expresses only one allele of the OR. In other words, ORs are expressed mono-allelically. The problem of determining which OR has been bound by odorant is reduced to distinguishing which olf. sensory neuron has been activated. Ressler; Vassar 1993 Chess 1994