* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download SNP Discovery by sequencing 1000 genomes
Genomic imprinting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Y chromosome wikipedia , lookup
Quantitative trait locus wikipedia , lookup
X-inactivation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Copy-number variation wikipedia , lookup
Population genetics wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Minimal genome wikipedia , lookup
Genealogical DNA test wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Oncogenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Frameshift mutation wikipedia , lookup
Genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Human genome wikipedia , lookup
Genome editing wikipedia , lookup
Public health genomics wikipedia , lookup
SNP genotyping wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Genome-wide association study wikipedia , lookup
Human Genome Project wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
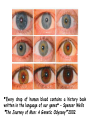
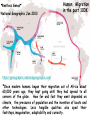
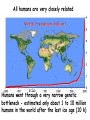
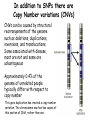
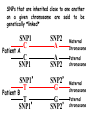
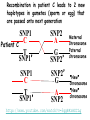
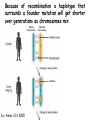
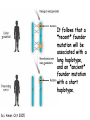
Applications in Bioinformatics, Proteomics, and Genomics SNPs (1) J. Gray (UT) [email protected] Science Dec 21 2007 The realization that DNA differs from person to person much more than researchers had suspected, may transform medicine but could also threaten personal privacy. www.sciencemag.org/sciext/btoy2007 Todays lecture 1: Understanding Genetic Variation 2. What are SNPs? 3. Why should we care about SNPs ? 4. SNP Discovery – The SNP Consortium/The International HapMap/The 1000 Genomes Project 5. Haplotypes and how chromosomal recombination gives rise to new Haplotypes 6. Overview of SNP detection methods 1: Understanding Human Genetic Variation “ Every drop of human blood contains a history book written in the language of our genes” - Spencer Wells “The Journey of Man: A Genetic Odyssey” 2002 Founder mutations Two men born in the US - thousands of miles apart - have a propensity to absorb iron so well that it can cause organ damage - a condition known as hereditary hemochromatosis. The error in their genes originated in a single European ancestor, whose ancestors now number nearly 22 million including the two men above (who might be surprised to know they are related). The original mutation is known as a “founder mutation”. The study of these mutations is intimately linked to the study of the recent evolution and spread of the human species. “Restless Genes” National Geographic Jan 2013 Human Migration in the past 100K https://genographic.nationalgeographic.com “ Once modern humans began their migration out of Africa about 60,000 years ago, they kept going until they had spread to all corners of the globe. How far and fast they went depended on climate, the pressures of population and the invention of boats and other technologies. Less tangible qualities also sped their footsteps,imagaination, adaptability and curiosity. All humans are very closely related Humans went through a very narrow genetic bottleneck - estimated only about 1 to 10 million humans in the world after the last ice age (10 k) Human demographic history has shaped the pattern of variation observed in modern populations. The general concensus is that Africa is the cradle of modern humans (approx 200k years ago) Genetic data shows the ALL non-Africans are the descendants of a small group of Africans that moved into the middle east about 70K yrs ago. The greatest diversity of genetic markers is in Africa indicating it was the earliest home of modern humans. Only a handful of people - carrying a few markers - left Africa seeding the genetic makeup of the rest of the world. National Geographic March 2006 See also http://www.bradshawfoundation.com/stephenoppenheimer/ Genetic mutations that act as markers, trace the journey of human migration. The earliest known mutation to spread outside of Africa is M168 (about 50 K yrs ago) This graphic shows the Y chromosome of a Native American man with various mutations including M168, proving his African ancestry. Founder mutations on Y chromosome give rise to Haplotypes “Eurasian Adam” In human genetics, Haplogroup CT is a Ychromosome haplogroup, defining one of the major lines of common ancestry of humanity along fatherto-son male lines. Men within this haplogroup have Y chromosomes with the SNP mutation M168, along with P9.1 and M294. These mutations are present in all modern human male lines except A and B, which are both found almost exclusively in Africa. Y-DNA Haplogroup Mutations Table The Y haplotype is very stable because there is no recombination happening with any other chromosome. The mitochondrial genome supplies a similar grouping in the maternal lineage. Haplogroups Mutations A no mutations B SRY10831.1 C SRY10831.1>M168 D SRY10831.1>M168>M174 E SRY10831.1>M168>M96 F SRY10831.1>M168>M89 G SRY10831.1>M168>M89>M201 H SRY10831.1>M168>M89>M69 I SRY10831.1>M168>M89>M170 J SRY10831.1>M168>M89>M304 K SRY10831.1>M168>M89>M9 L SRY10831.1>M168>M89>M9>M11 M SRY10831.1>M168>M89>M9>M5 N SRY10831.1>M168>M89>M9>M214 O SRY10831.1>M168>M89>M9>M214>M175 O3 SRY10831.1>M168>M89>M9>M214>M175>M122 P SRY10831.1>M168>M89>M9>M45 Q SRY10831.1>M168>M89>M9>M45>P36 R SRY10831.1>M168>M89>M9>M45>M207 R1b SRY10831.1>M168>M89>M9>M45>M207>M343 The pattern of genetic diversity in modern human populations, is the result of many evolutionary processes. New tools/resources promise to help identify functional mutations important for normal phenotypic variation as well as susceptibility to genetic disease. The same approaches are just as important for deciding how to protect biodiversity and in aiding plant breeding and animal husbandry Q: How much do humans differ ? A: very very very little! But everyone is unique Human genome project involved DNA from 9 individuals from diverse ethnic backgrounds Identified about 26,000-40,000 genes Also revealed about 1.5 million Single Nucleotide Polymorphisms – SNPs (Snips) These are the most prevalent form of genetic variation in humans – it was estimated there are 20 million SNPs (about 0.6% of the total genome) 2: What is a SNP ? (Single Nucleotide Polymorphism) 2: So what is a SNP ? GCATGCATGCATGCAT |||||||||||||||| CGTACGTACGTACGTA GCATGCAaGCATGCAT |||||||||||||||| CGTACGTtCGTACGTA Gene allele A1 Gene allele A2 Comparing DNA between two individuals shows that about every 1.5 kb there is one base pair difference – a single nucleotide polymorphism (SNP). When a variant nucleotide is present in more than one percent of a population, that DNA position is the location of the SNP. (less than 1% considered “rare” alleles). Only 93% 59% 39% 2% of genome encodes protein of all annotated genes have 1 SNP have 5 or more SNPs have 10 or more SNPs Often scientists distinguish between ancient “founder mutations” where surrounding DNA is same as others in the population and “hot spot mutations” which occur in error prone regions. Sci. Amer. Oct 2005 Old Originals versus numerous newcomers Sickle cell anemia is most often caused by a “founder mutation” Achondroplasia (a form of human dwarfism) ordinarily results from a “hotspot mutation” Sci. Amer. Oct 2005 Noteworthy Founder Mutations Gene Condition Mutation origin HFE Iron overload Possible Advantage of 1 copy Across Europe Protection from anemia CFTR Cystic fibrosis SW Europe Across Europe Protection from diarrhea HbS Sickle cell Africa To New World Protection from disease Middle East malaria ALDH2 Alcohol Far east Asia North & West Protection from toxicity across Asia alcoholism LCT Lactose Asia West & North Allows animal milk tolerance across Eurasia consumption GJB2 Deafness Middle East West & North Unknown across Europe FV Blood clots W. Europe Worldwide Protection from Leiden sepsis Sci. Amer. Oct 2005 NW Europe Migration In addition to SNPs there are Copy Number variations (CNVs) CNVs can be caused by structural rearrangements of the genome such as deletions, duplications, inversions, and translocations. Some associated with disease, most are not and some are advantageous Approximately 0.4% of the genome of unrelated people typically differ with respect to copy number This gene duplication has created a copy-number variation. The chromosome now has two copies of this section of DNA, rather than one. 3: Why should we care about SNPs ? 3: Why should we care about SNPs ? We want to know the basis of human variation and disease susceptibility How can some who never smoke get lung cancer and others who smoke heavily stay cancer free ? Why do some people exposed to HIV never develop AIDS ? SNPs are useful to....... 1: DNA fingerprinting for criminal or parental identification 2: Help map polygenic/disease traits by comparing DNA of groups with and without inheritance of that disease 3: Genotype-specific medication (pharmacogenomics) 4: Study human evolution 4: SNP Discovery 4: SNP Discovery The urgency and importance of identifying thousands of SNPs resulted in 11 major pharmaceutical and technology companies cooperating and one large scientific trust to underwrite the work – the TSC. Also see http://www.wtccc.org.uk Example of SNP Discovery The Whitehead Institute –isolated DNA from 10 ethnically diverse humans (Pilot phase). NA10965 NA10540 NA10470 NA08779A NA11322 NA11589 NA13820 NA13117 NA12615 NA11997 Ameridian Female Melanese Female Biaka Pygmy Male American Black Female Chinese Male Japanese Female Russian Male CEPH/Amish Female CEPH/French Male CEPH/Utah Female Example of SNP Discovery First a pool of 24 DNAs was digested with one of several restriction enzymes, size fractionated and cloned into M13-based vectors. Individual clones sequenced, repeats discarded, gene pairs accepted only if 99% homologous. SNPfinder algorithm used to find base pair discrepancies, repeated clusters removed Validations of SNPs – using Phred scores 20-51 Validation in 8 individuals – PCR and sequence candidate SNP regions – reject SNP if heterozygous in all individuals (assumed to be in a repeat region). Isolated more than 1.5 million SNPs www.hapmap.org See also http://www.ncbi.nlm.n ih.gov/SNP/ The Goal of the International HapMap Project is to develop a “haplotype” map of the human genome, the HapMap, which will describe the common (not rare) patterns of human DNA sequence variation (variants in >1% of population). The HapMap has become a key resource for researchers to use to find genes affecting health, disease, and responses to drugs and environmental factors. Phase 3 has been completed and there >6 million SNPs defined. The information is freely available. (see Nature 27 Oct 2005 for report on phase 1 of project, Nature 18 Oct 2007 for phase II and 2 Sep 2010 for phase III) Sequencing Entire Genomes – The Terabyte era July 10, 2008 DNA sequencing enters the terabase era The Wellcome Trust Sanger Institute announced something remarkable: its scientists had sequenced 300 human genomes in six months. In perspective. They sequenced more DNA every 2 seconds than was sequenced during the first five years of international genomesequencing efforts, from 1982 to 1987. The institute has now sequenced 1 trillion = 1000 billion letters of the genetic code. The cost of sequencing a human genome has fallen from $3 billion in 2001 (Human Genome Project) $1 million in 2007 (for James Watson) $50,000 in 2010 (James Lupski) Expect ~$1000 in 2013 (NIH goal) -Get used to it! Oxford Technolgies MiniION USB sequencer claims a $1000 genome Technology is not commercial yet High error rate Cannot provide a full genome (yet) Need to know Diploid genome https://www.nanoporetech.com Comments on limitations http://www.facebook.com/notes/brandon-colby-md/a-physiciansthoughts-on-oxford-nanopores-minion-and-gridion-dnasequencing-devi/320675544646237 SNP Discovery by sequencing individual genome Lupski, J.R. et al., New England Journal of Medicine 362:11811191 2010 James Lupski, a physician-scientist who suffers from a neurological disorder called Charcot-Marie-Tooth, searched for the genetic cause for > 25 years…….. Late last year, he finally found it-by sequencing his entire genome -in SH3TC2 (the SH3 domain and tetratricopeptide repeats 2 gene) – cost ~$50,000 First to show how whole-genome sequencing can be used to identify the genetic cause of an individual's disease. "I have hundreds of thousands of differences from all the other genomes that have been sequenced. I expect that to hold true for others. Everyone is truly unique.” SNP Discovery by sequencing family genomes How much genetic variation in each family? Sequenced entire genome of two parents and 2 children who both have a recessive genetic disease named Miller Syndrome Estimated a human intergeneration mutation rate of ~1.1 x 10-8 per position per haploid genome a high degree of certainty that each parent passes 30 new mutations—for a total of 60—to their offspring Also narrowed candidate genes to just four Roach et al., Analysis of Genetic Inheritance in a Family Quartet by Whole-Genome Sequencing. Science DOI: 10.1126/science.1186802 March 2010 SNP Discovery by sequencing 1000 genomes With advances in sequencing technology, the 1000 genomes project became feasible – revealed more SNPs than the HapMap project. www.genome.gov/27542240 - useful video tutorials Whose 1000 genomes? Deep whole-genome sequencing of trios (motherfather-daughter) from 2 populations Low-coverage sequencing of 179 unrelated individuals from 4 populations Exon sequencing of 906 randomly-selected genes in 697 individuals from 7 populations. Yielded 4.9 terabases of sequence! 15 million SNPs 1 million Indels (Insertions/Deletions) 20,000 structural variantsp SNP Discovery by sequencing 1000 genomes http://browser.1000genomes.org/index/html All data is deposited at 1000genomes.org Paper: A map of human variation from population-scale sequencing Nature Vol 467 p 1061 October 2010 What sequencing 1000 genomes reveals Variation is not evenly distributed in the genome (higher in telomeres, lower in gene dense regions Diversity in exons is half that of introns Most SNPs were already known of which 56% were present in all population panels, 25% in a single panel Of new SNPS (novel variants) 4% were found in all panels and 84% in only one (more rare variants) New germline mutations = about 1 in 10-8 68,300 novel non-synonymous variants About 340-400 Loss-of-function variants per individual, affecting 250-300 genes (we are all mutants!) Any individual genome differs by about 10,000 nonsynonymous variants from the ref sequence Culture cell lines accumulated hundreds of mutations not present in the germline Would you have your genome sequenced if you could afford it? Yes No Undecided 81% 9% 10% If you had your genome sequenced would you want to know everything? Yes No Undecided 74% 16% 10% In 2013 Researchers were able to identify 50 people whose DNA had been posted anonymously on the Internet for genetics studies. The results highlight a trade-off in making genetic data widely available for researchers and protecting personal privacy. 5: Haplotypes and how chromosomal recombination gives rise to new Haplotypes xyz XYZ xyz Xyz Xyz xYz xYZ During meiosis, homologous chromosomes (1 from each parent) pair along their lengths. The chromosomes cross over at points called chiasma. At each chiasma, the chromosomes break and rejoin, trading some of their genes. This recombination results in genetic variation (new haplotypes). Crossing over occurs during Meiosis http://www.youtube.com/watch?v=BhJf9MHHmc4 http://www.youtube.com/watch?v=3qgBKrAZCLg Crossing Over during Meiosis increases genetic variability http://www.dnatube.com/video/350/Crossi ng-Over-increases-genetic-variability If every homologous pair in humans has just one crossing over event then there will many possible new gametes (sperm or eggs) with many new haplotypes (depends on how the chromosomes randomly segregate and how many). SNPs that are inherited close to one another on a given chromosome are said to be genetically “linked” SNP1 C Patient A C SNP1 SNP2 A A SNP2 SNP1’ T Patient B T SNP1’ SNP2’ G G SNP2’ Maternal chromosome Paternal chromosome Maternal chromosome Paternal chromosome Haplotype refers to the set of alleles on one particular chromosome Patient C has two haplotypes SNP1 C Patient C T SNP1’ SNP2 A G SNP2’ Maternal chromosome Paternal chromosome Each haplotype is passed on to offspring as a complete unit unless recombination occurs between them to create new haplotypes A Trio is the genotype of mother father and offspring Recombination in patient C leads to 2 new haplotypes in gametes (sperm or egg) that are passed onto next generation SNP1 C Patient C T SNP1’ SNP2 A G SNP2’ Maternal chromosome Paternal chromosome SNP1 C T SNP1’ SNP2’ G A SNP2 “New” chromosome “New” chromosome http://www.youtube.com/watch?v=3qgBKrAZCLg Because of recombination a haplotype that surrounds a founder mutation will get shorter over generations as chromosomes mix Sci. Amer. Oct 2005 It follows that a “recent” founder mutation will be associated with a long haplotype, and an “ancient” founder mutation with a short haplotype. Sci. Amer. Oct 2005 6: How to detect SNPs ? SNP assay requirements a: Assay must be easily developed from sequence information b: Low cost of assay development (reagents/personnel) c: Assay must be robust d: Easily automated e: Simple analysis, accurate genotype calling f: Scalable assay (up to millions/day) g: Low cost per genotype assay Genotyping methods are evolving rapidly and costs greatly decreasing How can we detect SNPs ? Since most association studies require genotyping large numbers of individuals with a large number of SNPs then SNP assays must clearly distinguish between different alleles. there are several methods and this is an area of intense investigation and improvement………… Sequence-specific SNP Detection Methods 1: Hybridization: Allele-specific probes that only hybridize when there is a perfect match several methods to detect hybridization Affymetrix® SNP Array 6.0 1.8 million SNPs ~ $400 http://www.affymetrix.com/estore/browse/staticHtmlContentTemplate.jsp?stati cHtmlMediaId=m1621192&isHtmlStatic=true&navMode=35810&aId=productsNav Sequence-specific SNP Detection Methods 2: Nucleotide incorporation: addition of nucleotides with DNA polymerase can only occur if 3 ’ end of primer is a perfect match with SNP This method can be miniaturized and large numbers of SNPs assayed in a short time e.g. Illumina Infinium II Assay Protocol - can assay 650,000 SNPs on one chip - three day protocol from start to finish Now Infinium HD does up to 1.2 million www.illumina.com Illumina Omni 5 million SNPs $580 For online video see http://www.illumina.com/applications/genot yping.ilmn Next lecture 1: Mapping complex traits using SNPs 2: Genome Wide Association Studies (GWAS) 3. Example of complex trait mapping Using SNP analysis to find gene linked to retinal dystrophy