* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download The Use of Cell-Free Systems to Produce Proteins for Functional
Genetic code wikipedia , lookup
Endomembrane system wikipedia , lookup
Expanded genetic code wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Bottromycin wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Protein domain wikipedia , lookup
Metalloprotein wikipedia , lookup
Gene expression wikipedia , lookup
Protein folding wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Interactome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein purification wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
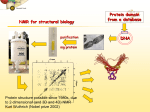
The Use of Cell-Free Systems to Produce Proteins for Functional and Structural Applications Julia Fletcher, Shiranthi Keppetipola, Ashley Getbehead, Federico Katzen and Wieslaw Kudlicki Invitrogen Corporation • 1600 Faraday Avenue • Carlsbad, California 92008 • USA Figure 1 – More functional protein Results Figure 3 – 100% incorporation of Selenomethionine into a membrane protein Figure 5 – Cell-free expression of SUMO for NMR analysis Se‐Met 188 98 62 98 62 49 49 38 28 •Reactions are easily scalable and flexible for expression of desired amounts of protein without the need for any special equipment. 38 EmrE EmrE 28 17 14 17 14 6 6 M L FT W E1 E2 E3 M L FT W E1 E2 E3 T FT W1 W2 E1 E2 E3 E4 M M 15N labeled‐ Sumo 15 Feed Reaction Incubate 30 min 1 ml Continue incubation up to 6 hrs Milligram quantities of protein 1 ml Figure 2 – Mg amounts of product 2 ml 100 90 80 70 60 50 40 30 20 10 0 13600 14299.03 EmrE Methionine 2480.1 14486.58 EmrE Seleno‐ methionine Figure 6 – N-HSQC spectra from crude and purified preparations of SUMO Purified •The Expressway™ Milligram system can be used to efficiently incorporate Selenomethinone for crystal structure analysis. •Expressway™‐NMR minimizes the amounts of labeled amino acids (1ml feed vs. 10ml dialysis) required for labeling due to smaller reaction volumes. •Expressway™‐NMR is supplied with a separate amino acid module allowing desired concentrations to be tailored as needed. 14200 14800 •Protein expression and labeling in the Expressway™‐NMR system provides protein samples sufficient for NMR analysis without the need for purification. 0 15400 Mass (m/z) Invitrogen’s Expressway™ Milligram and NMR systems use a patented buffer formulation of novel compounds to regenerate the diphospho‐ and monophospho‐nucleotides and maintain a pool of available NTPs for in vitro protein synthesis. These intermediates are supplied to the reaction through discrete “feeds”, in a very simple format, requiring no special equipment or devices to generate milligram levels of protein. These intermediates work in conjunction with an enhanced E. coli extract to drive higher yields of functional synthesized proteins (Figures 1 and 2). A single one‐milliliter reaction combined with a one‐milliliter feed can generate milligrams of soluble, biologically active protein. Reduced levels of amino acid scrambling make it possible to efficiently label the products with modified amino acids. The products are suitable for protein crystallization and NMR spectroscopy (Figures 3, 4, 5, and 6). The addition of a mix of detergents and liposomes to the reaction permitted, for the first time, the synthesis of milligram levels of an integral membrane protein suitable for crystallization (Figures 3 and 4). Finally, the Expressway™ NMR protein expression system provides protein samples of adequate concentration and purity for direct NMR analysis (Figure 6). Conclusions • Expressway™‐Milligram generates milligram quantities of proteins in a single reaction simply through the addition of feeding buffer during the reaction. kDa Met %Intensity Abstract Robust and dependable protein expression systems are the subject of increasing demands in the areas of genomics and proteomics, regardless whether studying single or multiple proteins from a functional or structural standpoint. Invitrogen’s Expressway™ systems allow the in vitro synthesis of recombinant proteins in a single reaction tube or in a multiplexed format in a just a few hours. We now report two additions to this product line, which are targeted to the structural proteomics field: Expressway™ Milligram and Expressway™ NMR. The core of these two kits consists of an improved buffer system and cell lysate that allows the synthesis of milligram amounts of protein in a 2‐ml‐ reaction format in just 3 hours using circular or linear DNA templates. The Expressway™ Milligram has been used to produce milligram amounts of otherwise difficult to express proteins such as membrane proteins. Functional and structural data will be presented. Expressway™ NMR provides an efficient and economical platform for stable‐isotope labeling of proteins. By employing this kit, protein purification is not longer needed to obtain clean and reliable HSQC‐spectra in under five hours. Figure 4 – Crystals and Diffraction pattern of a membrane protein synthesized in vitro References Crude Crystals of Selenomethionine‐labeled EmrE Diffraction from crystals (up to 3.8A) Pornillos, O.,Chen, Y. J.,Chen, A. P. and Chang, G., 2005. X‐ray structure of the EmrE multidrug transporter in complex with a substrate. Science. 310, 1950‐3. Keppetipola, S., Kudlicki, W., Nguyen, B. D., Meng, X ., Donovan, K. and Shaka AJ., 2005. From Gene to HSQC in Under Five Hours: High Throughput NMR Proteomics. JACS (submitted) [In collaboration with Scripps Research Institute (G Chang laboratory, Pornillos et al, 2005), Invitrogen’s Research Grant] [In collaboration with the AJ Shaka laboratory, UC Irvine] Expressway.Milligram Cell‐Free E. coli Expression System with pEXP5‐NT/TOPO® and pEXP5‐NT/TOPO® ,5 rxns, Cat No. K9900‐96 without vector, 5 rxns K9900‐97 Expressway.NMR Cell‐Free E. coli Expression Systemwith pEXP5‐NT/TOPO® and pEXP5‐ NT/TOPO®, 5 rxns, Cat No. K9900‐98, without vector, 5 rxns, Cat No. K9900‐99 Invitrogen Corporation • 1600 Faraday Avenue • Carlsbad, California 92008 USA • Telephone: 760 603 7200 • FAX: 760 602 6500 • Toll Free Telephone: 800 955 6288 • E-mail: [email protected] • www.invitrogen.com