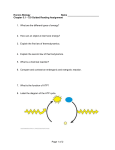
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 1 Confusion from last week: Purines and Pyrimidines
Genetic code wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Photosynthesis wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Microbial metabolism wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Gene expression wikipedia , lookup
Messenger RNA wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Proteolysis wikipedia , lookup
Metalloprotein wikipedia , lookup
Citric acid cycle wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Biosynthesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Biochemistry wikipedia , lookup
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 10: Energy & Enzymes http://compbio.uchsc.edu/hunter/bio5099 [email protected] Confusion from last week: “Small” RNAs are RNAs not involved in protein production, e.g. snRNA – Ribosomal rRNA is part of the machinery that produces proteins (more in a moment) First, a distinction without a difference. “Small RNA” is somewhat ambiguous term – Fine to say that rRNA is not a small RNA Second, mRNA and tRNA are representations of protein itself, where rRNA is not, hence the distinction... Purines and Pyrimidines Pyrimidines are single dinitrogen heterocycles Purines are two fused dinitrogen heterocycles 1 Back to the Ribosome Recall the Ribosome is the location of protein synthesis, where mRNA is translated. Today, we finish up by looking at tRNAs and the other molecules of the complex. tRNA function Transfer RNA (tRNA) translates individual codons to their corresponding amino acids. One end has an anti-codon which binds to the mRNA. The tRNA codon sequence is the same as the gene sequence – mRNA is inverse of DNA, tRNA is inverse of mRNA – RNA, so U instead of T Other end binds the appropriate amino acid. – Specifically, but not too strongly (has to release it!) tRNA structure tRNA has a fixed conformation (contrast with “floppy” mRNA ) RNA secondary structure is how nucleotides in the RNA bind to other nucleotides in it RNA tertiary structure is the complete 3D conformation. 2 Other parts of the Ribosome Initiation factors: – Proteins which start the process of translating mRNA Elongation factors: – Proteins which ensure that the translation proceeds appropriately. – E.g. Peptidase, which creates the peptide bonds Termination factors: – Detect STOP codons, and end the translation Example biomolecular study Ribosome is a good example of a biomolecular machine – Complex components, intricately assembled – Key function: the mechanism for creation of all proteins – Need to understand • • • • All of its components (their structures and functions) Their interactions Energetics (which we ignored so far). Synthesis, distribution, control, degradation.... Hierarchical study of structure and function Core Functions of life Energy: Capture, storage and use of energy Anabolism: Creation of biomolecules – Protein (Central Dogma, ribosomes, tRNAs) – DNA duplication and transcription – Lipids and energy storage molecules Catabolism: Breakdown of inputs – Getting energy from food, sunlight or redox potential – Creating material building blocks Control: Sensing and acting on state (of the self & of the environment) Reproduction: Making offspring 3 Molecular Power Adequate -G must be coupled to reactions that don't occur spontaneously (most of biology). – Too little energy, and necessary reactions don't occur – Too much energy, and bonds inside important molecules (e.g. proteins) can be disrupted, doing damage. Also, may need other forms of energy (redox) Where does the energy come from? How is it delivered when and where it is needed? Energy metabolism ATP: powering life Energy is obtained by the breakdown of food (or photosynthesis, or redox reactions) Energy is used by nearly all living processes Adenosine Triphosphate (ATP) is the main energycarrying molecule of life ATP Adenosine is the same as the nucleotide in RNA. Triphosphate is the addition of 3 PO4s Phophate bonds are “high energy” in that they release a lot of energy under hydrolysis Hydrolysis produces -G 7.3 Kc/mol 4 Amazing ATP Use of ATP is huge. – A cell uses about 12nM ATP/day (7.5X1015 molecules) – For a “basal” human, about 125 moles of ATP (>130 lbs) per day. Twice that for physically active person. When [ATP] > [ADP], - G can be 11–13 Kc/Mol Demand can change 10x in <0.001 second Reserves are tiny. Must be able to produce large quantities of ATP very quickly. – ATP is more like electricity than like a battery. Preview of ATP synthesis Breakdown of sugars produces two moles of ATP for each mole of sugar – Glycolysis Aerobic metabolism can use oxygen to create 36M ATP/M sugar! – Citric Acid or Tricarboxylic Acid (TCA) or Krebbs cycle. Various compounds which can be rapidly interconverted to ATP are used for storage – Lipids, Glycogen, Creatinine (more like batteries) More details when we get to catabolism... Reductive power Oxidation/Reduction is a necessary step in many biosynthetic reactions – E.g. making/breaking a double bond Requires chemical energy Potential carried by set of related compounds: – Nicotinamide Adenine Dinucleotide (NAD / NADH) • More often found in catabolism reactions – NAD phosphate / NADPH • More often found in anabolism reactions – Flavin Adenine Dinucleotide (FAD / FADH) 5 Coenzymes Molecules like ATP and NADH are “helpers” to many different kinds of enzymatic reactions Required cofactors are called “coenzymes” and are often “carriers” of functional groups: – Coenzyme A carries acetyl group – Biotin carries a carboxyl group (COO) Vitamins are often coenzyme precursors – Niacin (B3) is precursor to NAD – Riboflavin (B2) to FAD Also called “currency metabolites” Enzymes Enzyme activity (i.e. possible reactions) defines the fundamental chemical steps life has at its disposal (to turn food into babies). In order to understand metabolism, you need to have a sense of what enzymes can do. Nearly all enzymes are proteins, but some RNAs and DNAs also have enzymatic activity Classified by the type of reaction(s) catalyzed – Six broad groups, 3982 officially accepted classes Oxidoreductases Catalysts for any redox reaction Subclassed by donor and receptor molecules Commonly named (de-)hydrogenases and reductases Example: Alcohol Dehydrogenase – Oxidation of ethanol (also octanol, etc.) – Donor is CH-OH – Acceptor is NAD+ 6 Transferases Transfer a functional group Subclassed by what is transferred – E.g. one carbon, nitrogen-containing, phosphate group Example: Hexokinase. Adds a phosphate group to a sugar (glucose, manose, etc.) Uses both phosphate and energy from ATP. Hydrolases Special class of transferases in which water is the acceptor. Subclassed by the type of bond acted on (e.g. esters, peptide bonds, etc.) and substrates Example: Glucose 6-phosphotase, which undoes what hexokinase does: Glucose-6-phosphate + H20 Glucose + PO4 Lyases Cleavage of a bond by other than hydrolysis or oxidation. When acting on a single substrate, forms a new ring or double bond Subclassed on the bond to be broken Commonly named “...lase” or, when reverse reaction, “synthase” E.g. Pyruvate decarboxylase (double bond formed is in CO2). 7 Isomerases Catalyze changes within a molecule (no change in composition) Subclassed based on type of change e.g. chirality, cis- vs. trans-, intramolecular transfers, etc. Example: Glucose-6-phosphate isomerase, creates fructose6-phosphate, removing a carbon from a 6 membered ring Ligases Ligases are enzymes that catalyse the joining of two molecules Subclassed on type of bond formed Sometimes called “synthetase” Example: Glutamine synthetase, adds NH4 to Glutamate to make Glutamine Enzyme Kinetics How to calculate the concentrations of reactants and products with enzymes? Useful (if simplified): Michaelis-Menton – Substrate (= reactant other than enzyme) reversibly binds to enzyme, but transition to product is irreversible: E+S k-1 k1 ES kcat E+P – Assume [E] << [S], and, initially, [P] << [S] – E+S ES reaches equilibrium quickly, kcat is rate limiting – Let the velocity of reaction V = dP/dt = kcat[ES] 8 Michaelis-Menton (2) We can define V in terms of substrate concentration and parameters Km and Vmax Define Km = [E][S]/[ES] = (k-1+kcat)/k1 ≅ k-1/k1 Vmax when enzyme is saturated, [E]total= [ES] Km = [S] when V = ½ Vmax V= Vmax[S] Km + [S] V0 = Vmax[S](Km + [S]) Interpreting Enzyme Constants Vmax and Km different for every pair of enzymes and substrates 1/Km measures affinity of enzyme for substrate Kcat/Km (constant part of Vmax/Km) measures efficiency of enzyme in transforming substrate into product Enzyme summary Enzymes catalyze an amazing variety of reactions, but not everything is possible Many enzymatic reactions would be thermodynamically unfavorable, and therefore require coupling to -∆G (generally from ATP) Most enzymatic reactions can run in either direction but are used primarily in one Kinetics can be complex, but often first order (Michaelis-Menton) approximations are good 9