* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download apoptosis
Cell encapsulation wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Cell culture wikipedia , lookup
Cell membrane wikipedia , lookup
Cell growth wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell nucleus wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Endomembrane system wikipedia , lookup
Cytokinesis wikipedia , lookup
Signal transduction wikipedia , lookup
List of types of proteins wikipedia , lookup
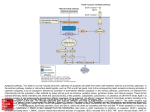
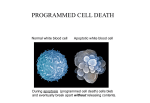
BB20023:DNA: make, break & disease 2004-05 APOPTOSIS There are two kinds of cell death, apoptotic (programmed cell death) and necrotic. NECROSIS APOPTOSIS Morphological features loss of membrane integrity Membrane blebbing, no loss of integrity swelling of cytoplasm & mitochondria Aggregation of chromatin at nuclear membrane total cell lysis Shrinking of cytoplasm & nuclear condensation no vesicle formation Fragmentation into smaller bodies disintegration of organelles Formation of apoptotic bodies Mitochondria become leaky Biochemical features Loss of regulation of ion homeostasis Energy dependent No energy requirement Non random DNA fragmentation Random digestion of DNA Prelytic DNA fragmentation Postlytic DNA fragmentaion Activation of caspase cascade Tightly regulated process Release of various factors into cytoplasm Physiological features Affects groups of cells Affects individual cells Evoked by non physiological disturbances Induced by physiological stimuli Phagocytosis by adjacent cells Phagocytosis by macrophages Significant inflammatory response No inflammatory response Caspases: the central executioners: (cysteinyl aspartate specific proteases) These are highly conserved proteases that usually exist as inactive zymogens that are activated to induce cell death. So far, at least 14 homologues have been identified, 11 in humans. Caspases can be broadly divided into either 1) Initiator caspases: Caspases 2, 8,9,10 or 2) Effector caspases: caspase 3,6,7 Structure: Caspases comprise of 3 domains a highly variable NH2 domain large subunit (~20kD) small subunit ( ~10kD) 2 key features: variable N domain regulates activation all domains derived from proenzyme at cleavage specific sites Highly specific unusual & absolute requirement for cleavage after aspartic acid recognition of at least 4 amino acids NH2 terminals to the cleavage site How do caspases disassemble a cell? It slices, it dices, and that's not all! Selectively cleave a discrete set of proteins in several ways Inactivate inhibitors of apoptosis eg bcl-2, or CAD/ICAD Direct disassembly of the cell structures e.g. nuclear lamina. Indirect cleaving of proteins involved in cytoskeletal regulation eg. Gelsolin Dr. MV Hejmadi BB20023:DNA: make, break & disease 2004-05 How are caspases activated? More than one way to skin a cat Mechanisms of caspase activation include proteolytic cleavage by an upstream caspase (a), induced proximity (b) and holoenzyme formation (c). Proteolytic cleavage by an upstream caspase is straightforward and effective, and is used mostly for activation of downstream, effector caspases. It is probably also used for induction of apoptosis by noncaspase proteases, such as granzyme B. In the second mechanism, recruitment or aggregation of multiple procaspase-8 molecules into close proximity somehow results in cross-activation. The actual process is most probably more sophisticated and more tightly regulated than shown in panel b. In holoenzyme formation, cytochrome c and ATP-dependent oligomerization of Apaf-1 allows recruitment of procaspase-9 into the apoptosome complex. Activation of caspase-9 is mediated by means of conformational change, not proteolysis. Stoichiometry of the apoptosome is not known; it is shown in panel c as a hexamer solely for aesthetic reasons The roads to ruin: A) Caenorhabditis elegans: One of the apoptotic pathways is triggered by internal signals of the CED system CED-3 & 4 promote apoptosis CED-9 inhibits apoptosis Apoptotic stimuli causes CED-9 dissociation by EGL-1 thereby activating CED-3. B) Mammalian cells: two major apoptotic pathways 1) External signals Specific ‘death ligands’ Loss of contact with surroundings 2) Internal signals Irreparable internal damage Conflicting signals for cell division Dr. MV Hejmadi BB20023:DNA: make, break & disease 2004-05 External signal driven by death receptors (DR) e.g. CD95 (or Fas/Apo) Each CD95L trimer binds to 3 CD95 leading to DD clustering. FADD ( Fas associated death domain/ Mort 1) binds via its own DD FADD effector DD to analogous domain in caspase –8 Caspase –8 oligomerisation drives activation thru self cleavage Caspase –8 then activates downstream effector caspases like caspase –9 (CED-9 homolog) Apoptosis initiation B) Internal signal driven by inactivation of bcl-2, structural & functional homolog of CED-9. One model for Apaf-1 regulation by the bcl-2 family. Bcl-xL binds toApaf-1 death signal provokes interaction of Bik/ Bax with Bcl-xL, Apaf-1 bind to procaspase-9 (in presence of cytochrome c) promote dimerisation & activation by autocatalysis. Caspase –9 subsequently activates effector caspases leading to apoptosis References: 1) Science (1998) Vol 281: No 5381; pgs 1298-1326 2) Nature (2000) 12th Oct, Vol 407 Insight articles a. The biochemistry of apoptosis by MO Hengartner pp 770 - 776 3) Molecular mechanisms of caspase regulation by S Reidl and Y Shi. Nature Reviews in Mol & Cell Biology (Nov 2004) Vol 5 pp 897 4) Apoptosis: Molecular mechanisms by Wu, Min; Ding, Han–Fei and Fisher, David E www.els.net Dr. MV Hejmadi