* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Ch 4 - Tacoma Community College
Survey
Document related concepts
Tissue engineering wikipedia , lookup
Cell growth wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell culture wikipedia , lookup
Signal transduction wikipedia , lookup
Cell encapsulation wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cell nucleus wikipedia , lookup
Cytokinesis wikipedia , lookup
Cell membrane wikipedia , lookup
Transcript
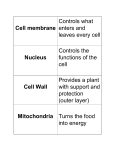
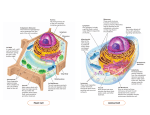
Ch 4 Tour of the Cell Microscopic Worlds • Microscopes led to the discovery of the cell – Light microscopes – – – – Cell membrane - yes Large macromolecules - no Microtubules - no Mitochondria - maybe just barely – Many bacteria - yes Microscopic Worlds Light microscope Scanning Electron Differential Inference-contrast Transmission Electron Cell Size • House DNA, protein molecules and internal structures • Obtain nutrients and diffuse nutrients and O2 • Smaller cells have a greater surface area to volume ratio than do larger cells – Surface area is significant for diffusion and osmosis Surface area : Volume 10 m 30 m 30 m Surface area of one large cube 5,400 m2 10 m Total surface area of 27 small cubes 16,200 m2 • Volume= 30 um *30 um* 30 um=27000um • SA (large)= 6*(30um*30um)=5,400 um • SA (small)=(6*(10um*10um))*27=16,200 um Domains of Life • The 3 domains of life – Bacteria (prokaryotic cells) – Archaea (prokaryotic cells) – Eukarya (all other life forms) Cells • Prokaryotic • Eukaryotic – Protists, fungi, plants, animals Colorized TEM 15,000 – Bacteria & Archaea Prokaryotic cell Nucleoid region Nucleus Eukaryotic cell Organelles Prokaryotic cells are simpler & usually smaller than Eukaryotic cells In Common • • • • Bounded by plasma membrane Ribosomes Cytoplasm DNA as genetic material Prokaryote • Do not have membrane bound nucleus • Have a cell wall outside their plasma membrane • Circular DNA strands • No membrane bound organelles Prokaryotic flagella Ribosomes Capsule Cell wall Plasma membrane Nucleoid region (DNA) Pili Eukaryote Rough endoplasmic reticulum Smooth endoplasmic reticulum • Membrane bound nucleus • Linear DNA • Membrane bound organelles Nucleus Flagellum Not in most Lysosome plant cells Centriole Peroxisome Microtubule Intermediate Cytoskeleton filament Microfilament Ribosomes Golgi apparatus Plasma membrane Mitochondrion Eukaryotic Cells • A typical animal cell: • Contains a variety of membranous organelles (underlined) Rough endoplasmic reticulum Smooth endoplasmic reticulum Nucleus Flagellum Not in most plant cells Lysosome Ribosomes Centriole Golgi apparatus Peroxisome Microtubule Cytoskeleton Plasma membrane Intermediate filament Mitochondrion Figure 4.4A Microfilament Eukaryotic Cells Rough endoplasmic reticulum • A typical plant cell NUCLEUS Golgi apparatus Ribosomes Smooth endoplasmic reticulum CYTOSKELETON: Central vacuole Microtubule Chloroplast Intermediate filament Cell wall Plasmodesmata Microfilament Mitochondrion Peroxisome Plasma membrane Cell wall of adjacent cell Categories of Organelles • Structural support, movement, communication – Cytoskeleton, plasma membrane, cell wall (plants) • Manufacturing – Nucleus, ribosomes, endoplasmic reticulum, Golgi apparatus • Energy processing – Mitochondria (animal), chloroplasts (plants) • Hydrolysis – Lysosomes (animals), vacuoles (plants), peroxisomes Plasma Membrane • Forms boundary around cell • Controls and regulates material transport -Semi permeable • Phospholipid bilayer Plants and Cell Walls • Cell wall – Cellulose • Connect by plasmodesmata – Channels between adj. cells Walls of two adjacent plant cells Vacuole Plasmodesmata Layers of one plant cell wall Cytoplasm Plasma membrane Cytoskeleton • Cell’s internal skeleton – Helps organize structure and activities – Consists of network of protein fibers Tubulin subunit Actin subunit Fibrous subunits 25 nm 7 nm Microfilament 10 nm Intermediate filament Microtubule Cytoskeleton • Microfilaments (actin filiments) – Enable cells to change shape and move • Intermediate filaments – Reinforce the cell and anchor certain organelles • Microtubules – give the cell rigidity, provide anchors for organelles, act as tracks for organelle movement Tubulin subunit Actin subunit Fibrous subunits 7 nm Microfilament 25 nm 10 nm Intermediate filament Microtubule Nucleus Chromatin Nucleolus Pore Nucleus • Contains most of the Two membranes cells DNA of nuclear envelope • Eukaryotic chromosomes made of chromatin • Enclosed by nuclear enveloper • Nucleolus Rough endoplasmic reticulum Ribosomes – rRNA synthesized Ribosomes • Free and bound ribosomes • Composed of 2 subunits – Involved in protein synthesis Endomembrane System • Interconnected structurally and functionally – Physically connected Rough ER OR – Connected via vesicles Transport vesicle from ER to Golgi Transport vesicle from Golgi to plasma membrane Plasma membrane Nucleus Vacuole Lysosome Smooth ER Nuclear envelope Golgi apparatus Rough Endoplasmic Reticulum • Membrane continuous with nuclear envelope Rough ER Nuclear envelope • Bound ribosomes – Produces proteins – Transported or secreted Figure 4.7 Ribosomes Smooth ER Rough ER TEM 45,000 – Makes more membrane – Transferred via vesicles Smooth ER Fig. 4-9b Transport vesicle buds off 4 Ribosome Secretory protein inside transport vesicle 3 Sugar chain 1 2 Glycoprotein Polypeptide Rough ER Smooth ER • Lacks bound ribosomes • Involved in metabolic processes – Synthesis of lipids, hormones, enzymes • Stores calcium Golgi Apparatus • Vesicles from ER go to Golgi • Receives and modifies products “Receiving” side of Golgi apparatus Golgi apparatus Golgi apparatus New vesicle forming “Shipping” side of Golgi apparatus Transport vesicle from the Golgi TEM 130,000 Transport vesicle from ER Mitochondria • Cellular respiration Outer membrane Inner membrane Cristae Matrix TEM 44,880 – Converts chemical energy to ATP Mitochondrion – Phospholipid bilayer membrane – Has own DNA and Intermembrane space ribosomes Chloroplasts • Convert solar energy to chemical energy (photosynthesis) • Stroma – Contains DNA, ribosomes and enzymes • Thylakoids – Interconnected sacs that form stacks called granum Endosymbosis • Hypothesis of endosymbosis – Mitochondria and chloroplasts were once small prokaryotes living independently – At some point, began living within larger cells Lysosomes • Digestive enzymes enclosed by membrane sac • Destroy ingested bacteria, recycle damaged Lysosome organelles • Break down food Digestion Vesicle containing damaged mitochondrion Lysosome Digestive enzymes Plasma membrane Digestion Food vacuole Lysosomes Rough ER Transport vesicle (containing inactive hydrolytic enzymes) Golgi apparatus Plasma membrane “Food” Engulfment of particle Lysosomes Food vacuole Figure 4.10A Digestion Lysosome engulfing damaged organelle Vacuoles • Membranous sac • Central Vacuole Nucleus – hydrolytic function Chloroplast • Contractile • Food Colorized TEM 8,700 Central vacuole Perixosomes • Metabolize fatty acids • Enzymes that digest peroxides • Come only from ER • Cilia Movement – Short appendage, numerous • Flagella – Long appendage • Microtubules wrapped in plasma membrane – Anchored by basal body • Moves via bending Figure 4.17A LM 600 Colorized SEM 4,100 Movement Figure 4.17B Extracellular matrix • Holds cells to tissues • Protect and support PM – Integrins • Regulate behavior, transmit information, coordinate cells Cell Junctions •Tight junctions can bind cells together into leakproof sheets •Anchoring junctions link animal cells into strong tissues •Gap junctions allow substances to flow from cell to cell Tight junctions Anchoring junction Gap junctions Extracellular matrix Space between cells Figure 4.18B Plasma membranes of adjacent cells Fig. 4-23