* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Advanced Techniques in Molecular Biology
Protein (nutrient) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gene regulatory network wikipedia , lookup
Protein moonlighting wikipedia , lookup
History of molecular evolution wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Point mutation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Molecular neuroscience wikipedia , lookup
Biochemistry wikipedia , lookup
Western blot wikipedia , lookup
Expression vector wikipedia , lookup
Protein adsorption wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Gene expression wikipedia , lookup
Biosynthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Proteolysis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
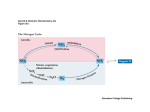
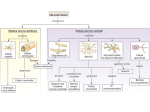
Advanced Techniques in Molecular Biology Sonchita Bagchi [email protected] Some basic Molecular Biology techniques • • • • • • DNA cloning Polymerase Chain Reaction Gel electrophoresis molecular hybridization Mutagenesis DNA and protein arrays Model organism Tissues attained and processed – parafin sections, cryo sections National Human Genome Research Institute (NHGRI) http://www.genome.gov/12514471 Ref: European Molecular Biology Laboratory: Explore Model organisms at EMBL Detection techniques •Fluorophore or fluorochrome •3,3'-diaminobenzidine tetrahydrochloride (DAB) •AminoEthyl Carbazole (AEC) •Alkaline Phosphatase (AP) http://www.immunohistochemistry.us/index.php?page=ihc-principle Quantum dots Quantum dots shine brighter, last longer, and are often less toxic than dyes previously used. In addition, specific organic molecules can be attached to the quantum dots for tracking. Advantages over conventional fluorophores •Photo stability •Narrow spectra Source: Kim Krieger, Science 2004 Quantum dots are semiconductive crystals that have been synthesized to be as small as possible, normally from 2-30 nanometers wide. Quantum dots can be synthesized so accurately that they can be tuned to absorb and release specific energy amounts. Source:Ishikawa-Ankerhold et.al. Molecules 2012 Functional studies using Co-immunoprecipitation Western Blot with antibody against the protein (if known!) Advantages: • Proteins that interact in a typical Co-IP are post-translationally modified and conformationally natural. • In Co-IP, proteins interact in a non-denaturing condition which is almost physiological. Disadvantages: • The signals of low-affinity of protein interactions might not be detected. • There might be a third protein in certain protein-protein interaction. • To choose an appropriate antibody, the target protein needs to be properly predicted. Or there would not be a positive result in Co-IP. • WB can be difficult to achieve. Proximity Ligation Assay (PLA) Source:Duolink Fluorescence Resonance Energy Transfer The energy is transferred from an excited donor fluorochrome to another molecule or acceptor without emission of a photon. FRET relies on the close physical interaction of the two molecules less than approximately 10 nm apart. Thus, FRET can be used to determine molecular interaction/molecular proximity beyond the resolution limits of the classical light microscope. Source:Life Technologies •Molecules must be <10nm apart. •Donor emission must overlap Acceptor excitation Fluorescence recovery after photobleaching (FRAP) Source: University of Gothenburg, Centre for cellular imaging RNA interference Source: National Institute of General Medical Sciences Overexpression of genes •Constitutive promoters •Inducible promoters Source: Muller et. al. Cytokine, 2008 Fluorescence Microscopy Confocal Microscopy Electron Microscopy Atomic force microscopy (AFM) Source: Syddansk Universitet Source: Bagchi et. al. Molecular Microbiology, 2008 Source: Lehenkari et. al. Ultramicroscopy, 2000 Questions?? Advanced Techniques in Molecular Biology Part 2 Epigenetics The term ‘Epigenetics’ was coined by Conrad H. Waddington in 1942, which was derived from the Greek word “epigenesis” which originally described the influence of genetic processes on development. Genetic assimilation (1990) is a process by which a phenotype originally produced in response to an environmental condition, later becomes genetically encoded via artificial selection or natural selection. Today “Epigenetics” refers to covalent modification of DNA, protein, or RNA, resulting in changes to the function and/or regulation of these molecules, without altering their primary sequences. Epigenetics is the reason why a skin cell looks different from a brain cell or a muscle cell. All three cells contain the same DNA, but their genes are expressed differently, which creates the different cell types. Is it possible to pass epigenetic changes to future generations? • DNA methylation • Histone modification • Non-coding RNA (ncRNA)-associated gene silencing DNA methylation: the addition of a methyl group to part of the DNA molecule, which prevents certain genes from being expressed. Determination of differential methylation with methylation-sensitive and methylation-insensitive restriction enzymes. Genome-wide methods to measure DNA methylation Histone Modifications Histone modifications are proposed to affect chromosome function. The first mechanism suggests modifications may alter the electrostatic charge of the histone resulting in a structural change in histones or their binding to DNA. The second mechanism proposes that these modifications are binding sites for protein recognition modules that recognize acetylated or methylated amino acid. Posttranslational modifications of histones create an epigenetic mechanism for the regulation of a variety of normal and disease-related processes. Purifying Histones Non-coding RNA (ncRNA)-associated gene silencing • Chromatin remodelling • Transcriptional regulation • Post-transcriptional regulation Molecular phylogenetics Phylogenetic systematics deals with identifying and understanding evolutionary relationships among the many different kinds of life on earth, both living (extant) and dead (extinct). • Alignment—building the data model and extracting a dataset. • Determining the substitution model—consider sequence variation. • Tree building. • Tree evaluation. Protein Modeling or Molecular Modeling • Identify the proteins with known three-dimensional structures that are related to the target sequence. • Align the related three-dimensional structures with the target sequence and determine those structures that will be used as templates. • Construct a model for the target sequence based on its alignment with the template structure(s). • Evaluate the model against a variety of criteria to determine if it is satisfactory. Prion - proteinaceous infectious particle • Prion is an abnormal form of a normally harmless protein found in the brain that is responsible for a variety of fatal neurodegenerative diseases of both animals and humans. • Prions can enter the brain through infection, or they can arise from mutations in the gene that encodes the protein. Once present in the brain, prions multiply by inducing benign proteins to refold into the abnormal shape. • The normal protein structure is thought to consist of a number of flexible coils called alpha helices. In the prion protein some of these helices are stretched into flat structures called beta strands. • The normal protein conformation can be degraded rather easily by cellular enzymes called proteases, but the prion protein shape is more resistant to this enzymatic activity. Thus, as prion proteins multiply they are not broken down by proteases and instead accumulate within nerve cells, destroying them. Glutamine-Glutamate cycle in depression, mood disorder and schizophrenia Histological analysis of SLC38A6 (SNAT6) expression in mouse brain shows selective expression in excitatory neurons with high expression in the synapses. Publications so far Histological analysis of SLC38A6 (SNAT6) expression in mouse brain shows selective expression in excitatory neurons with high expression in the synapses. Bagchi S, Baomar HA, Al-Walai S, Al-Sadi S, Fredriksson R. PLoS One. 2014 Apr; 9(4): e95438-e95438. The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. Sundberg BE, Wååg E, Jacobsson JA, Stephansson O, Rumaks J, Svirskis S, Alsiö J, Roman E, Ebendal T, Klusa V, Fredriksson R. J Mol Neurosci. 2008 Jun; 35(2):179-93. SLC38A6 is widely expressed in the mouse brain The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. Sundberg BE, Wååg E, Jacobsson JA, Stephansson O, Rumaks J, Svirskis S, Alsiö J, Roman E, Ebendal T, Klusa V, Fredriksson R. J Mol Neurosci. 2008 Jun; 35(2):179-93. SNAT6 is selectively expressed in excitatory neurons SNAT6 has high expression in the synapses Genes predicted to influence SNAT6: According to the PCR results the top three genes interacting with SLC38A6 are as follows: 1. PAG (GLS1) (glutaminase) [already investigated] 2. CTPS2 (CTP Synthase) and it Catalyses the formation of CTP from UTP with the concomitant deamination of glutamine to glutamate. ATP+UTP+GLUTAMINE→ADP+Pi+CTP+GLUTAMATE 3. GRM2 (Glutamate Receptor, Metabotropic): It is a metabotropic glutamate receptor that appears on excitatory neurons. It is worth mentioning that according to the computational simulation, two other slc38 transporters seem to be interacting with A6 and they are SLC38A1 and SLC38A5. The results shows Gls, Grm-2 ,ctps2 and Dner had the biggest relative fold change. It is interesting to note that GLS and Grm2 are upregulated in the wild type, while Dner and Ctps2 are upregulated in the siRNA treated cells. Functional basis: Speculations! • Gls2 is an phosphate-activated amidohydrolase that catalyzes the hydrolysis of glutamine to glutamate and ammonia. This protein (primarily expressed in the brain and kidney) plays an essential role in generating energy for metabolism, synthesizing the brain neurotransmitter glutamate and maintaining acid-base balance in the kidney. Thus it makes sense that in case of decreasing the SNAT6( siRNA condition), the production of the Gls protein decreases, as there is not enough glutamine to hydrolyse to glutamate. • Grm-2 (Metabotropic glutamate receptor 2) exists on the surface of neurons and they receive glutamate. As the glutamine percentage is decreasing in the treated cells, the need for more glutamate decreases and thus they are down regulated. • One of the main functions of Dner is clearing of glutamate, thus it seems very reasonable to increase its production in order to compensate the loss of glutamine. • ctps2 is a rate-limiting enzyme in the synthesis of cytosine nucleotides (ATP + UTP + glutamine → ADP + Pi + CTP + glutamate). Hence, ctps2 is upregulated as the cell tries to decrease the production of glutamate to conserve glutamine. Direct Interaction between SNAT6 and CTPase2 • PLA (raised in same species) • CoIP (too low Antibody conc.) Questions??