* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download doc BIOL202-16
SNP genotyping wikipedia , lookup
DNA polymerase wikipedia , lookup
Primary transcript wikipedia , lookup
Metagenomics wikipedia , lookup
Gene therapy wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Genome evolution wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genealogical DNA test wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA vaccination wikipedia , lookup
Genome editing wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Designer baby wikipedia , lookup
Genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Molecular cloning wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Microevolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
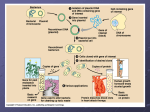
BIOL 202-date:2/08 Lecture 16 Cloning: generating recombinant (hybrid) DNA molecules o The way to do that is to cut the DNA into parts with restriction enzymes. And place them into vectors. o The restriction fragments from donor DNA are not all the same size. By digesting the insert DNA, you can anneal the complementary strand and ligase can close the breaks between strands. o The goal is not to isolate the vector, but to get a huge quantity of that DNA insert. o o All these fragments are inserted into the same vectors but generating different recombinant vectors. o o Then by using electrical-chemical process we can transform the bacteria with the recombinant vector, while bacteria genome can’t get out. o o The bacteria will then use it’s DNA replication machinery to create multiple copies of the recombinant vectors before replication, cell division would subsequently produce more bacteria that can replicate the recombinant DNA. o Many copies of the same recombinant DNA inside the same bacteria. Selecting plasmids with DNA insertion. o How do you know that you have an insert inside the plasmid? pBR322 is the first vector used by biologist. This vector has an origin of replication, ampicilin resistant gene, in both of those genes you have many unique restriction enzyme sites, meaning that they exist only once inside the bacteria. By inserting a fragment gene inside one of those plasmid genes, then that gene will be completely destroyed. pUC18 vector is a derivative of pBR322 vector, it also have a origin of replication and ampicilin resistant gene, this gene is used to select for transformed bacteria, meaning bacteria that have taken up the plasmid, thus be able to synthesize ampicilin resistant proteins, unlike untransformed bacteria. pUC18 vector also have a laz gene (different) than pUC18, a pUC18 vector that has not underwent successful recombination will have the lac Z gene intact because the gene of interest was not inserted into the lac Z gene. In the presence of the dye X-gal, transformed bacteria with a pUC18 vector that have a functional Lac Z gene (did not underwent recombination) will produce a blue colored dye. Bacteria transformed with the desired gene of interest will have a disrupted Lac Z gene, and therefore unable to produce the blue color when exposed to X-gal. A polylinker is a sequence of DNA inserted into vectors o Clones containing DNA fragment will form colonies. o o We plate the bacteria on ampicilin/agar/x-gal plate in a way that would produce individual colonies at low density o At first we won’t see anything, but after 1 to 2 days, visible colonies of each clone will form. o Bacterial cells divide every 20-30 minutes o Each colony represents a clone of the corresponding single cell. o All the colonies we see are transformed bacteria that incorporated the plasmid. o The white colonies contains plasmid with a functional ampicilin resistance gene but a malfunctional Lac Z gene, this means that plasmid has an inserted gene, however we can’t be sure that the inserted gene contains our YMWG. (your most wanted gene) o In the blue colonies, X-gal is transformed into a blue dye. o Will not be used to sequence the human DNA because we can only use up to 10kb for one fragment, and the human genome have 3 million base pairs. o Cloning will not give you 100% transformed bacteria, nor 100% success rate in recombination of vectors. We used phage λ, which is a virus that infects E.coli. The phage contains 48 kilo base pairs, this allows 10-20kb to be inserted into the DNA. o Digest genomic DNA with a restriction enzyme, by doing that we can insert these fragments inside the λ phage, by isolating the left and right arm. o We have to generate a partial fragments of the donor DNA, because small fragments will not fit inside the λ phage. o We use fragments of about 15kb, we can isolate this DNA of this length by using electrophoresis and cutting out gel containing DNA fragments from 12-20kb. o o Then use ligase with this extracted DNA and the left and right arm of the λ phage. o The result is a long chain of tandem recombinant DNA units This is way too big to fit into the head of the virus. o The virus will recognize a sequence at the end of left and right arm, anything that is in between these sequence “cos” site will be able to fit inside the virus only if the DNA insert is between 15-20kb. This means that the total sequence length, including left and right arm will have to be around 45-50kb. o The DNA that can be inserted inside a λ phage is twice as long as that inserted inside plasmid pUC18. o o The bacteria is infected by the virus, and plaques are produced when contents of infected bacteria is spilled out to infect neighboring bacteria. Note that no ampilicin is added. o However, this insert is still comparatively small, o Cos sites determined what fits in the head of the virus. o People put cos sites at the end of a phage λ DNA, this will generate a plasmid, when the 2 cos sites attach itself, called a cosmids. o With cosmids, the DNA insert can be around 35-45kb. o The bigger the DNA insert, the less fragments you will have to analyze. In cosmids, the virus does not have gene required for infection, instead a normal plasmids will result inside the bacteria. you will need ampr genes and origin implication. BAC (bacterial artificial chromosome)-bigger vector o Even bigger fragment insert can be used with BAC bacteria (150-300kb) Inserting recombinant DNA into bacteria o o in method A, we are using a plasmid to transform the bacteria. The vector contains an ampicilin resistant gene. o In method B, we are using a phage to insert a linear piece of DNA with COS ends. Once inserted inside the bacteria, the phage DNA will form a cosmid, this method is called transduction. We will need a selectable marker, like in A. o In method C, we are using bacteriophage to infect bacteria and analyzing the clones inside progeny phages that are released by lysis and detected as plaques on agar plates. We don’t need a selectable marker. o o