* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Topic 14: Protein Synthesis
Ancestral sequence reconstruction wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Western blot wikipedia , lookup
Polyadenylation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Protein adsorption wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
List of types of proteins wikipedia , lookup
Messenger RNA wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein structure prediction wikipedia , lookup
Non-coding RNA wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene expression wikipedia , lookup
Biochemistry wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Bottromycin wikipedia , lookup
Expanded genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
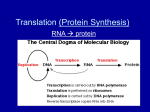
TOPIC 14: PROTEIN SYNTHESIS (lecture 23) OBJECTIVES: 1. Understand the overall structure and role of tRNA’s in translation. 2. Be able to describe the various parts of the ribosome and the roles of the P site and A site in translation; understand the overall process of initiation, elongation and termination of transcription. 3. Know the major kinds of post-translational modifications that take place after proteins are synthesized. 4. Know what a point mutation is, how point mutations can be produced, and be able to differentiate between silent, missense, nonsense and frame shift mutations Translation: The synthesis of protein. (fig. 17.12, overview) The mRNA carries a faithful record of the amino acid sequence of the protein as specified by the gene sequence. How is this used to make protein? fig. 17.13- transfer RNA (tRNA); specialized RNA molecules that literally are involved in transferring the appropriate amino acid to the growing polypeptide chain 1. roughly 80 nucleotides long 2. at the 3’ end in a site where a particular amino acid will be attached 3. consists of three loops; the middle of which corresponds to a site known as the anticodon site; it has base sequence that is complementary to codons on the mRNA 4. there are 41 different tRNA’s ; there are 61 different codons so some tRNA’s can bind more than one codon. There is a relaxation of the base-pairing rules for the third position of the condon; this is called wobble. a. U can pair with either A or G when in the third position b. if the nucleotide Inosine (I) is present, it can bind to U,C and A in the third position of the codon c. see fig. 17.4; note that codons coding for the same amino acid typically only differ in the nucleotide in the third position aminoacylsynthetases- these are enzymes which attach a particular amino acid to the 3’ end of the corresponding t-RNA; they are specific for each amino acid (see fig. 17.14); note: ATP is hydrolyzed here! ribosomes (fig. 17.15) - RNA and protein complex; site of protein synthesis; the RNA is known as ribosomal RNA (rRNA) and consists of large and small subunits with three binding sites - P site, A site and mRNA binding site. Fig. 17.16- atomic structure of ribosome complex 1 Fig. 17.12- overview of the process of translation; binding of aminoacyl tRNA to mRNA followed by peptide formation and then translocation and binding of another aminoacyl tRNA. Initiation of translation (fig. 17.17)1. small rRNA subunit binds to a region of the mRNA near the beginning of the region coding for the polypeptide chain 2. an initiator tRNA with the anticodon for methonine (UAC) binds to the AUG (“start”) site 3. the large rRNA subunit attaches in such a way that the initiator tRNA is in the P site and the A site is unoccupied 4. proteins called initiation factors are involved in this process; GTP is hydrolyzed Elongation (fig. 17.18)1. binding of complementary anticodon of a tRNA to the codon in the A site 2. peptide bond formation 3. the tRNA in the P site is released and the ribosome advances one codon downstream so that the A site is again unoccupied; this means that the peptidyltRNA is now positioned in the P site 4. another complementary tRNA then binds to the codon in the A site…. Termination (fig. 17.19)1. when the termination codon reaches the A site, elongation will stop 2. a release factor binds to the codon and causes a water molecule to be added to the polypeptide chain causing the polypeptide to be ultimately released. Polyribosomes (fig. 17.20)- at any given point in time, many ribosomes may be attached to a single message After synthesis the polypeptide will fold and assume its native 3-D structure; this is sometimes assisted by other proteins known as chaperonins. In addition, if it is a multiple subunit protein, subunits must come together to make the quaternary structure. Post-translational chemical modifications may take place: 1. cleavage by specific proteases; for instance, most mitochondrial proteins are synthesized in the cytoplasm. They have a leader peptide in the N-terminal region which allows the mitochondrion to recognize it and transport it into the matrix. The leader peptide is then clipped off and the protein is trapped in the mitochondrion. 2. covalent attachment of other molecules onto particular amino acid residuesphosphate, sugars, lipids 3. removal of N-terminal methionine 2 Signal sequences- like mitochondrial proteins, other proteins have signal sequences which target them for particular places in the cell; see fig. 17.21 for targeting mechanism into Golgi system. Mutation- an alteration in the genetic information in a cell; mutations can take place by large scale changes in DNA that might take place during gamete formation or cell division involving pieces of chromosomes or by point mutations (point mutation- a change in a single nucleotide in a gene). Large scale changes in genes and chromosomes- these mutations can involve drastic changes in which genes or portions of genes or portions of entire chromosomes are deleted or moved around to different places. Point mutations occur naturally during DNA replication; they are rare. However, certain agents such as ionizing radiation (X-rays etc), UV light and a variety of chemicals may produce mutations. These agents are known as mutagens. Ionizing radiation like X-rays and gamma rays- these cause the production of highly reactive chemical compounds known as free radicals; free radicals can break the phosphodiester bonds of DNA UV light- causes the formation of thymidine-thymidine dimers (adjacent T’s become covalently attached); presence will block DNA replication Chemical mutagens- may covalently add new carbons to nucleotides causing mispairing Kinds of point muations (fig. 17.24 a & b): 1. base pair substitution- one nucleotide is replaced by another; can have no effect on the amino acid sequence (silent); it can change the amino acid (missense) or it can put an inappropriate stop signal into the sequence (nonsense) 2. insertion or deletion- alters the reading frame of the message and can have catastrophic effects; these are known as frame shift mutations. Fig. 17.23- sickle cell anemia is the result of a single nucleotide substitution which results in a protein have a Val instead of Glu! This substitution produces profound consequences for the functioning of red blood cells. Fig. 17.25- Summary of transcription/translation 3