* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download File E-Leraning : METABOLISME
Fatty acid synthesis wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Butyric acid wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Biosynthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Blood sugar level wikipedia , lookup
Metalloprotein wikipedia , lookup
Phosphorylation wikipedia , lookup
Photosynthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Electron transport chain wikipedia , lookup
Microbial metabolism wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Biochemistry wikipedia , lookup
Citric acid cycle wikipedia , lookup
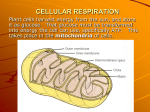
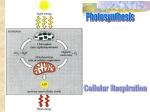
Enzim merupakan senyawa organik bermolekul besar yang berfungsi untuk mempercepat jalannya reaksi metabolisme di dalam tubuh tumbuhan tanpa mempengaruhi keseimbangan reaksi Enzim tidak ikut bereaksi, struktur enzim tidak berubah baik sebelum dan sesudah reaksi tetap Enzim sebagai biokatalisator Bagian enzim yang aktif adalah sisi aktif dari enzim Enzim diberi nama sesuai dengan nama substrat dan reaksi yang dikatalisis Biasanya ditambah akhiran ase Enzim dibagi ke dalam 7 golongan besar Klas Oksidoreduktase (nitrat reduktase) Transferase (Kinase) Hidrolase (protease, lipase, amilase) Tipe reaksi memisahkan dan menambahkan elektron atau hidrogen memindahkan gugus senyawa kimia Liase (fumarase) membentuk ikatan rangkap dengan melepaskan satu gugus kimia Isomerase (epimerase) Ligase/sintetase (tiokinase) Polimerase (tiokinase) mengkatalisir perubahan isomer memutuskan ikatan kimia dengan penambahan air menggabungkan dua molekul yang disertai dengan hidrolisis ATP menggabungkan monomer-monomer sehingga terbentuk polimer Komponen utama enzim adalah protein Protein yang sifatnya fungsional, bukan protein struktural Tidak semua protein bertindak sebagai enzim Enzim terdiri dari 1 ata lebih rantai polipeptida dan bagian yang bukan protein yang penting untuk aktifitas katalitik (kofaktor) Kofaktor: 1. gugus prostetik 2. koenzim 3. ion metal Gugus prostetik: senyawa organik yang mengikat kuat pada apoenzim Koenzim: senyawa organik yang berasosiasi secara tidak permanen dengan apoenzim, biasanya berupa vitamin. Beberapa contoh enzim yang memerlukan kofaktor 1. 2. 3. 4. 5. 6. NAD (koenzim 1) NADP (koenzim 2) FMN dan FAD Cytokrom: cytokrom a, a3, b, b6, c, dan f Plastoquinon, plastosianin, feredoksin ATP: senyawa organik berenergi tinggi, mengandung 3 gugus P dan adenin ribose Enzim dibentuk dalam protoplasma sel Enzim beraktifitas di dalam sel tempat sintesisnya (disebut endoenzim) maupun di tempat yang lain diluar tempat sintesisnya (disebut eksoenzim) Sebagian besar enzim bersifat endoenzim 1. 2. 3. 4. 5. 6. 7. 8. Enzim bersifat koloid, luas permukaan besar, bersifat hidrofil Dapat bereaksi dengan senyawa asam maupun basa, kation maupun anion Enzim sangat peka terhadap faktor-faktor yang menyebabkan denaturasi protein misalnya suhu, pH dll Enzim dapat dipacu maupun dihambat aktifitasnya Enzim merupakan biokatalisator yang dalam jumlah sedikit memacu laju reaksi tanpa merubah keseimbangan reaksi Enzim tidak ikut terlibat dalam reaksi, struktur enzim tetap baik sebelum maupun setelah reaksi berlangsung Enzim bermolekul besar Enzim bersifat khas/spesifik Suhu: optimum 300C, minimum 0 0C, maksimum 400C Logam, memacu aktifitas enzim: Mg, Mn, Co, Fe Logam berat, menghambat aktivitas enzim: Pb, Cu, Zn, Cd, Ag pH, tergantung pada jenis enzimnya (pepsin aktif kondisi masam, amilase kondisi netral, tripsin kondisi basa) Konsentrasi substrat, substrat yang banyak mula-mula memacu aktifitas enzim, tetapi kemudian menghambat karena: penumpukan produk (feed back effect) Konsentrasi enzim, peningkatan konsentrasi enzim memacu aktifitasnya Air, memacu aktifitas enzim Vitamin, memacu aktifitas enzim Penghambatan aktifitas enzim : 1. Kompetitif 2. Non kompetitif 3. unkompetitif Penghambatan kompetitif Penghambatan nonkompetitif Penghambatan unkompetitif Respirasi Heterotrof • Heterotrof membutuhkan molekul organik sebagai makanan. • Energinya diperoleh dari ikatan kimia di dalam molekul makanan seperti karbohidrat, lemak dan protein. Autotrof • Autotrof merupakan organisme yang dapat menggunakan sumber energi dasar (sinar matahari) untuk membuat energi. Mis: tumbuhan dan beberapa mikroorganisme • 2 Types – Photosynthetic autotrophs – Chemosynthetic autotrophs Chemosynthesis • Chemosynthesis is a process used by prokaryotic organisms to use inorganic chemical reactions as a source of energy to make larger organic molecules. Aerobic Vs. Anaerobic • Aerobic respiration requires oxygen. • Anaerobic respiration does not require oxygen. Aerobic Respiration • Aerobic cellular respiration is a specific series of enzyme controlled chemical reactions in which oxygen is involved in the breakdown of glucose into carbondioxide and water. • The chemical-bond energy is released in the form of ATP. • Sugar + Oxygen carbon dioxide + water + energy (ATP) Summary: 3 steps: 1st glycolysis 2nd Krebs cycle 3rd Electron Transport Chain (ETC) A Road Map for Cellular Respiration Cytosol Mitochondrion High-energy electrons carried mainly by NADH High-energy electrons carried by NADH Glycolysis Glucose 2 Pyruvic acid Krebs Cycle Electron Transport Figure 6.7 Glycolysis Stage 1: splitting glucose molecule (C6) into two PGALD (C3). 2 ATPs are used per glucose metabolism. • Stage 2: catalyzes the oxidation of PGALD to pyrvate. Generate s 4 ATPs per glucose metabolism. glucose ATP Glycolysis Hexokinase ADP glucose-6-phosphate Phosphoglucose Isomerase fructose-6-phosphate ATP Phosphofructokinase ADP fructose-1,6-bisphosphate Aldolase glyceraldehyde-3-phosphate + dihydroxyacetone-phosphate Triosephosphate Isomerase Glycolysis continued glyceraldehyde-3-phosphate NAD+ + Pi Glyceraldehyde-3-phosphate Dehydrogenase NADH + H+ Glycolysis continued. Recall that there are 2 GAP per glucose. 1,3-bisphosphoglycerate ADP Phosphoglycerate Kinase ATP 3-phosphoglycerate Phosphoglycerate Mutase 2-phosphoglycerate Enolase H2O phosphoenolpyruvate ADP Pyruvate Kinase ATP pyruvate 6 CH2OH 5 H 4 OH O H OH H 2 3 H OH glucose 6 CH OPO 2 2 3 ATP ADP H H 1 OH 5 4 Mg2+ OH O H OH 3 H 2 H 1 OH Hexokinase H OH glucose-6-phosphate 1. Hexokinase catalyzes: Glucose + ATP glucose-6-P + ADP The reaction involves nucleophilic attack of the C6 hydroxyl O of glucose on P of the terminal phosphate of ATP. ATP binds to the enzyme as a complex with Mg++. 6 CH OPO 2 2 3 5 H 4 OH O H OH 3 H H 2 OH H 1 OH 6 CH OPO 2 2 3 1CH2OH O 5 H H 4 OH HO 2 3 OH H Phosphoglucose Isomerase glucose-6-phosphate fructose-6-phosphate 2. Phosphoglucose Isomerase catalyzes: glucose-6-P (aldose) fructose-6-P (ketose) The isomerization is an electron shift where 2 electron from C2 carbon reduce the C1 aldehyde of the glucose6-phosphte molecule to an alcohol. Phosphofructokinase 6CH OPO 2 2 3 O 5 H H 4 OH 6CH OPO 2 2 3 1CH2OH O ATP ADP HO 2 3 OH H fructose-6-phosphate 5 Mg2+ 1CH2OPO32 H H 4 OH HO 2 3 OH H fructose-1,6-bisphosphate 3. Phosphofructokinase catalyzes: fructose-6-P + ATP fructose-1,6-bisP + ADP This highly spontaneous reaction has a mechanism similar to that of Hexokinase. The Phosphofructokinase reaction is the ratelimiting step of Glycolysis. . 2 1CH2OPO3 2C O HO 3C H 4C H H 5 C H Aldolase 2 CH OPO 2 3 3 OH 2C OH 1CH2OH 2 CH OPO 2 3 6 fructose-1,6bisphosphate O + O 1C H 2C OH 2 CH OPO 3 2 3 dihydroxyacetone glyceraldehyde-3phosphate phosphate Triosephosphate Isomerase 4.Aldolase catalyzes: fructose-1,6-bisphosphate dihydroxyacetone-P + glyceraldehyde-3-P The reaction is an aldol cleavage, the reverse of an aldol condensation. Note that C atoms are renumbered in products of Aldolase. 2 1CH2OPO3 2C O HO 3C H 4C H H 5 C H Aldolase 2 CH OPO 2 3 3 OH 2C OH 1CH2OH 2 CH OPO 2 3 6 fructose-1,6bisphosphate O + O 1C H 2C OH 2 CH OPO 3 2 3 dihydroxyacetone glyceraldehyde-3phosphate phosphate Triosephosphate Isomerase 5. Triose Phosphate Isomerase (TIM) catalyzes: dihydroxyacetone-P glyceraldehyde-3-P The dihidroxyacetone-P is isomerized to PGALD, thus this stage produce 2 molecules PGAL pe 1 glucose Triosephosphate Isomerase H H C OH C O + + H H CH2OPO32 dihydroxyacetone phosphate H OH H H C C + + H OH CH2OPO32 enediol intermediate O C H C OH CH2OPO32 glyceraldehyde3-phosphate The ketose/aldose conversion involves acid/base catalysis, and is thought to proceed via an enediol intermediate, as with Phosphoglucose Isomerase. Active site Glu and His residues are thought to extract and donate protons during catalysis. Glyceraldehyde-3-phosphate Dehydrogenase H 1C O OPO32 + H+ O NAD+ NADH 1 C + Pi H 2C OH H 2 C OH 2 CH OPO 2 3 3 glyceraldehyde3-phosphate 2 CH OPO 2 3 3 1,3-bisphosphoglycerate 6. Glyceraldehyde-3-phosphate Dehydrogenase catalyzes: glyceraldehyde-3-P + NAD+ + Pi 1,3-bisphosphoglycerate + NADH + H+ Glyceraldehyde-3-phosphate Dehydrogenase H 1C O OPO32 + H+ O NAD+ NADH 1 C + Pi H 2C OH H 2 C OH 2 CH OPO 2 3 3 glyceraldehyde3-phosphate 2 CH OPO 2 3 3 1,3-bisphosphoglycerate Exergonic oxidation of the aldehyde in glyceraldehyde- 3phosphate, to a carboxylic acid, drives formation of an acyl phosphate, a "high energy" bond (~P). This is the only step in Glycolysis in which NAD+ is reduced to NADH. Phosphoglycerate Kinase O OPO32 ADP ATP O O 1C H 2C OH 2 3 CH2OPO3 1,3-bisphosphoglycerate C 1 2+ Mg H 2C OH 2 3 CH2OPO3 3-phosphoglycerate 7. Phosphoglycerate Kinase catalyzes: 1,3-bisphosphoglycerate + ADP 3-phosphoglycerate + ATP This phosphate transfer is reversible (low DG), since one ~P bond is cleaved & another synthesized. The enzyme undergoes substrate-induced conformational change similar to that of Hexokinase. Phosphoglycerate Mutase O O C 1 O O C 1 H 2C OH 2 3 CH2OPO3 H 2C OPO32 3 CH2OH 3-phosphoglycerate 2-phosphoglycerate 8. Phosphoglycerate Mutase catalyzes: 3-phosphoglycerate 2-phosphoglycerate Phosphate is shifted from the OH on C3 to the OH on C2. Enolase O O C 1 H 2 C OPO32 3 CH2OH H O O C C OH O O C 1 OPO32 CH2OH 2C OPO32 3 CH2 2-phosphoglycerate enolate intermediate phosphoenolpyruvate 9. Enolase catalyzes: 2-phosphoglycerate phosphoenolpyruvate + H2O O O Pyruvate Kinase C 1 C 2 ADP ATP O O C 1 OPO32 3 CH2 phosphoenolpyruvate C 2 O 3 CH3 pyruvate 10. Pyruvate Kinase catalyzes: phosphoenolpyruvate + ADP pyruvate + ATP Krebs Cycle • Makes ATP • Makes NADH • Makes FADH2 Conversion of pyruvate to Acetyl CoA NAD+ NADH O HSCoA O H3C O pyruvate dehydrogenase complex pyruvate • 2 per glucose (all of Kreb’s) • Oxidative decarboxylation • Makes NADH CO2 O SCoA H3C acetyl CoA Net From Kreb’s • Oxidative process – 3 NADH – FADH2 – GTP • X 2 per glucose – 6 NADH – 2 FADH2 – 2 GTP • All ultimately turned into ATP (oxidative phosphorylation…later) Citrate Synthase Reaction (First) Aconitase Reaction O HO O O C CH2 C C CH2 C O O HO O O citrate aconitase O C CH HC C CH2 C O O O O isocitrate • Forms isocitrate • Goes through alkene intermediate (cis-aconitate) – elimination then addition • Hydroxyl moved and changed from tertiary to secondary – (can be oxidized) • 13.3kJ Isocitrate Dehydrogenase O HO C CH HC C CH2 C O O O O NADH CO2 O O isocitrate • • • • NAD O C C O CH2 CH2 C isocitrate dehydrogenase O O alpha ketoglutarate All dehydrogenase reactions make NADH or FADH2 Oxidative decarboxylation -20.9kJ Energy from increased entropy in gas formation α-ketoglutarate dehydrogenase O O C C O CH2 CH2 SCoA CoASH CO2 NAD C CH2 CH2 C NADH C O O alpha ketoglutarate O O O dehydrogenase alpha ketoglutarate • • Same as pyruvate dehydrogenase reaction Formation of thioester – endergonic – driven by loss of CO2 • increases entropy • exergonic • -33.5kJ succinyl CoA Succinyl CoA synthetase SCoA O C CH2 CH2 C O O succinyl CoA O GDP GTP CoASH succinyl CoA synthetase • Hydrolysis of thioester • Coupled to synthesis of GTP • -2.9kJ – Releases CoASH – Exergonic – Endergonic – GTP very similar to ATP and interconverted later O C CH2 CH2 C O O succinate Succinate dehydrogenase O O O C CH2 CH2 C O O FAD FADH2 succinyl CoA dehydrogenase C C H H C C O succinate • • Dehydrogenation Uses FAD – NAD used to oxidize oxygen-containing groups • Aldehydes • alcohols – FAD used to oxidize C-C bonds – 0kJ O O fumarate Fumarase O C C H H C C O O O O H2O fumarase O C HC OH CH2 C O fumarate • Addition of water to a double bond • -3.8kJ O malate Malate Dehydrogenase O O C HC OH CH2 C O O malate • • • • O NADH NAD malate dehydrogenase O O C C O CH2 C O oxaloacetate Oxidation of secondary alcohol to ketone Makes NADH Regenerates oxaloacetate for another round 29.7 kJ Net From Kreb’s • Oxidative process – 3 NADH – FADH2 – GTP • X 2 per glucose – 6 NADH – 2 FADH2 – 2 GTP • All ultimately turned into ATP (oxidative phosphorylation…later) INHIBITOR OF TCA CYCLE 1. Fluoroacetate • Condensation with CoA Fluoroacetyl-CoA • Condensation FluoroacetylCoA with Oxaloacetate Fluorocitrate inhibit Akonitase enzyme accumulation of citrate • Fluoroacetate pesticide 2. Malonate suksinat dehydrogenase enzyme 3. Arsenite α-ketoglutarate dehydrogenase enzyme REGULATION OF TCA CYCLE REGULATION OF TCA CYCLE Electron Transport System • The electrons released from glycolysis and the Krebs cycle are carried to the electrontransport system (ETS) by NADH and FADH2. • The electrons are transferred through a series of oxidation-reduction reactions until they are ultimately accepted by oxygen atoms forming oxygen ions. • 32 molecules of ATP are produced. What is the electron transport chain? A chain of protein complexes embedded in the inner mitochondrial membrane. Transports electrons and pumps hydrogen ions into the intermembrane space to create a gradient. Components of the electron transport chain (ETC) NADH dehydrogenase Ubiquinone (Q) Succinate dehydrogenase Cytochrome b-c1 Cytochrome c Cytochrome oxidase Arranged in order of increasing electronegativity (weakest to strongest) NADH produced from glycolysis VS VS NADH produced from Krebs cycle • NADH produced in the cytoplasm by glycolysis: CAN diffuse from outer mitochondrial membrane to intermembrane space. CANNOT diffuse from inner membrane to matrix. • NADH produced from Krebs cycle: Already in the matrix. What to do? • Cytosolic NADH (NADH that are produced by glycolysis) pass electrons to shuttles • Glycerol-phosphate shuttle: Transfers electrons from cytosolic NADH to FAD to produce FADH2 • Aspartate shuttle: Transfers electrons from cytosolic NADH to NAD+ to produce NADH Complex I: NADH dehydrogenase • NADH dehydrogenase oxides NADH back to NAD+ NADH + H+ NAD+ + 2H+ + 2e• The electrons are transferred to flavin mononucleotide (FMN) which reduces to FMNH2 FMN + 2H+ + 2e- FMNH2 • The electrons are then passed to iron-sulphur proteins (FeS) located in NADH dehydrogenase Complex I Continued… • Electrons are accepted by Fe3+ which is reduced to Fe2+ 2Fe3+ + 2e- 2Fe2+ • These two electrons are then given to ubiquinone (Q) • The two hydrogen ions are pumped into the intermembrane space *One hydrogen ion is pumped per electron transferred Ubiquinone (Q) • A mobile electron carrier 2Fe2+ gives 2e- to Q Q is reduced to QH2 (ubiquinol) and 2Fe2+ is oxidized 2Fe3+ • Carries electrons to complex III, cytochrome b-c1 Interactive Animation: http://www.brookscole.com/chemistry_d/ templates/student_resources/shared_re sources/animations/oxidative/oxidativep hosphorylation.html Complex II: Succinate Dehydrogenase • oxidation of succinate from Krebs cycle to fumarate Succinate + FAD Fumarate + FADH2 FADH2 then tries to oxidize back into FAD by passing its electrons to 2 Fe2+ 2Fe3+ + 2e- 2Fe2+ Complex II Continued... • These electrons from 2Fe2+ are then stolen by ubiquinone (Q), which carries them to complex III. • 2+ 3+ Q + 2Fe 2Fe + QH2 • Unlike complex I, complex II doesn’t have enough free energy for active transport of the hydrogen ions (protons) from FADH2 across the intermembrane . This is why oxidation of FADH2 only yields 2 ATP molecules instead of 3 ATP molecules like NADH. Complex III: cytochrome b-c1 • Contains cytochrome b, cytochrome c1, and FeS proteins. • QH2 passes two electrons to cytochrome b causing Fe3+ to reduce to Fe2+ • Same way from cyt b to FeS protein and then to cyt c1 Cytochrome C (c) • I’m a protein and like Q... • I am also a water soluble mobile electron carrier. I transport electrons one at a time from complex III to complex IV. Complex IV: Cytochrome Oxidase (The End of the Line) • This is the end of the line for electrons from NADH or succinate. • Oxygen (breathed in) is reduced and when combined with these electrons, they form... WATER ½ O2 (g) + 2H+ + 2e- H2O (l) THIS REACTION IS EXPLOSIVE! It’s highly exergonic, releasing large amounts of energy. Electrochemical gradient • A concentration gradient created by pumping ions into a space surrounded by a membrane that is impermeable to the ions. • Two components: electrical and chemical • Proton-motive force (PMF) moves protons through an ATPase complex on account of the free energy stored in the form of an electrochemical gradient of protons across a biological membrane. Chemiosmosis • Peter Mitchell – Nobel Prize in Chemistry in 1978 • A process for synthesizing ATP using the energy of an electrochemical gradient and the ATP synthase enzyme. • “osmosis” • After chemiosmosis, ATP molecules are transported through both mitochondrial membranes. Connections! • Electrons from NADH and FADH2 were passed down the ETC • As the electrons move down, energy released moves protons to create electrochemical gradient • Protons move through proton channels, and release energy to synthesize ATP from ADP and Pi • The many processes of ATP synthesis are all continuous Fermentation • Fermentation describes anaerobic pathways that oxidize glucose to produce ATP. • An organic molecule is the ultimate electron acceptor as opposed to oxygen. • Fermentation often begins with glycolysis to produce pyruvic acid. Anaerobic Cellular Respiration • Anaerobic respiration does not require oxygen as the final electron acceptor. • Some organisms do not have the necessary enzymes to carry out the Krebs cycle and ETS. • Many prokaryotic organisms fall into this category. • Yeast is a eukaryotic organism that performs anaerobic respiration. • First step in anaerobic respiration is also glycolysis Diagram Anaerobic Respiration Cytoplasm C6H12O6 glucose Alcoholic fermentation Bacteria, Yeast 2 ATP glycolysis Aerobic Respiration Lactic acid fermentation Muscle cells 2 ATP Krebs Cycle ETC Mitochondria Alcoholic Fermentation • Alcoholic fermentation is the anaerobic pathway followed by yeast cells when oxygen is not present • Pyruvic acid is converted to ethanol and carbon-dioxide. • 4 ATPS are generated from this process, but glycolysis costs 2 ATPs yielding a net gain of 2 ATPs. •Alcoholic fermentation—occurs in bacteria and yeast Process used in the baking and brewing industry—yeast produces CO2 gas during fermentation to make dough rise and give bread its holes glucose ethyl alcohol + carbon dioxide + 2 ATP ALCOHOL FERMENTATION Lactic Acid Fermentation • In Lactic acid fermentation, the pyruvic acid from glycolysis is converted to lactic acid. • The entire process yields a net gain of 2 ATP molecules per glucose molecule. • The lactic acid waste products from these types of anaerobic bacteria are used to make fermented dairy products such as yogurt, sour cream, and cheese. • Lactic acid fermentation—occurs in muscle cells Lactic acid is produced in the muscles during rapid exercise when the body cannot supply enough oxygen to the tissues—causes burning sensation in muscles glucose lactic acid + carbon dioxide + 2 ATP LACTIC ACID FERMENTATION PHOTOSYNTHESIS photosynthesis • Photo artinya cahaya dan sintesis artinya membuat • Proses di dalam tanaman yang mengubah carbon dioksida dan air menjadi gula dan oksigen • Terjadi di dalam kloroplas Photosynthesis: 6 CO2 + 6 H2O carbon dioxide + water = C6 H12 O6 + 6 O2 sugar + oxygen photosynthetic products often stored as starch •Starch = glucose polymer Tracking atoms STARCH Carbon Fixation • Penggabungan karbon dioksida menjadi senyawa organik disebut juga fiksasi karbon. • Fiksasi karbon terjadi selama reaksi yang tidak bergantung adanya cahaya/ siklus calvin/ reaksi gelap. Fig. 10.20 Calvin Cycle – Reminder of Steps! Karbon dioksida berdifusi masuk kedalam stroma kloroplas. Enzim , RibuloseBifosfatase ( Rubisco) memungkinkan karbon dioksida untuk bergabung dengan gula berkarbon 5 (Ribulose biphosphate). Senyawa beratom karbon 6 tersebut sifatnya UNSTABLE dan dipecah menjadi 2 gula berkarbon 3 (3-PGA) 2 3-PGA kemudian diubah menjadi G3P 1 G3P akan menjadi glukosa Lainnya diregenerasi kembali menjadi RibuloseBiphosphatase. Fig. 10.17 Types of photosynthesis • C3 – Kebanyakan tanaman • C4 – CO2 untuk sementara disimpan dalam bentuk asam organik 4-C. – Berguna: penyinaran yang lama, suhu tinggi, kurang CO2 – Rerumputan dan tanaman pertanian(e.g., jagung, sorgum, tebu) •CAM •Stomata terbuka pada malam hari •Berguna dalam kondisi iklim kering •Kebanyakan kaktusan (e.g., cacti, euphorbs, bromeliades, agaves) C3! • Siklus calvin dikenal sebagai lintasan C3 PGA • Contoh tumbuhan C3: C3 Plants • stomata TERBUKA pada siang hari dan TERTUTUP pada malam hari. • Ketika stomata terbuka, karbon dioksida dapat masuk dan Oksigen dapat keluar. • Pada saat cuaca panas, kering dan kehilangan air(transpirasi) menjadi MASALAH. Problem • Ketika stomata tertutup, karbon dioksida tidak dapat masuk ke dalam daun dan oksigen tidak dapat keluar dari daun. • Oksigen berkompetesi dengan karbon dioksida di dalam siklus calvin. • Ketika terlau banyak oksigen di dalam daun, oksigen akan berikatan dengan RUBISCO (ribulose biphosphase) ini disebut sebagai fotorespirasi. Competing Reactions • Rubisco dapat bereaksi dengan CO2 (Carboxylase Reaction) – bagus untuk produksi glukosa CH2OPO32- C O H C OH H C OH CH2OPO32Ribulose 1,5-bisphosphate CO2 O- O C H C OH CH2OPO323-Phosphoglycerate O- O C H C OH CH2OPO 323-Phosphoglycerate Competing Reactions • Rubisco dapat juga bereaksi dengan O2 (Oxygenase Reaction) – Tidak bagus untuk produksi glukosa CH2OPO32C O H C OH H C OH CH2OPO32Ribulose 1,5-bisphosphate O2 O- O CH2OPO32- C H C OH CH2OPO323-Phosphoglycerate C O O- Phosphoglycolate • Ketika oksigen berikatan dengan Rubisco kemudan akan berkombinasi dengan gula berkarbon 5 dan dirombak. • Siklus Calvin terhenti: gula berkarbon 5 tidak bisa menjadi gula dan dirombak menjadi air dan karbon dioksida. • Rubisco bereaksi dengan O2 dibanding CO2 • Terjadi dlam kondisi: – Konsentrasi oksigen tinggi – Kondisi panas • Fotorespirasi dipetkirakan menurunkan efisiensi fotosintesis sampai25%. Solusi Photorespiration Alternative Pathways Pada kondisi panas dan kering, tumbuhan harus melakukan lintasan alternatif. C4 dan CAM Fig. 10.21 C4 Pathway • Pada lintasan C4 fotosintesis terjadi pada mesofil dan selubung berkas pembuluh. – Reaksi terang di mesofil – Siklus calvin di Selubung berkas pembuluh Image taken without permission from C4 Pathway • CO2 difikasi menjadi molekul berkarbon 4 • Enzim tambahan– PEP Carboxylase that initially traps CO2 instead of Rubisco– makes a 4 carbon intermediate C4 Pathway • The 4 carbon intermediate “smuggles” CO2 into the bundle sheath cell • The bundle sheath cell is not very permeable to CO2 • CO2 is released from the 4C molecule goes through the Calvin Cycle C3 Pathway How does the C4 Pathway limit photorespiration? • Bundle sheath cells are far from the surface– less O2 access • PEP Carboxylase doesn’t have an affinity for O2 allows plant to collect a lot of CO2 and concentrate it in the bundle sheath cells (where Rubisco is) CAM Pathway • Fix CO2 at night and store as a 4 carbon molecule • Keep stomates closed during day to prevent water loss • Same general process as C4 Pathway • Has the same leaf anatomy as C3 plants How does the CAM Pathway limit photorespiration? • Collects CO2 at night so that it can be more concentrated during the day • Plant can still do the calvin cycle during the day without losing water Fig. 10.22 Sintesis Sukrosa dan Pati Synthesis of Starch and Sucrose Photosynthetic cell • CO2 chloroplast Sukrosa: produk penting dari fotosintesis – PGA RuBP 1,3 bisPGA • Cadangan gula yg penting – starch – Triose P sucrose • tap root of carrots and sugar beet (up to 20% dry weight) and in leaves, eg 25% leaf dry weight in ivy Bentuk utama translokasi C – – RuBP = ribulose 1,5-bis-phosphate (pentose) 3-PGA = 3-phosphoglycerate 1,3 bisPGA = 1,3 bis-phosphoglycerate accounts for most of CO2 absorbed from photosynthetic leaves (source leaves) in germinating seedlings after starch or lipid breakdown Sugar Translocation is Essential • Gula diperlukan dalam proses metabolisme – all the time, in all tissues • Sugars produced only – by source tissues • Translokasi terjadi pada: – source to sink over short term – from storage tissues to young tissues over long term Sugar composition of phloem sap • > 500 different species (100 families) of dicots (Zimmermann & Ziegler, 1975) Most families Aceraceae (maple) Anacardiaceae (cashew) Asteraceae (aster) Betulaceae (birch) Buddleiaceae (butterfly bush) Caprifoliaceae (honeysuckle) Combretaceae (white mangrove) Fabaceae (legume) Fagaceae (beech & oak) Moraceae (fig) Oleaceae (olive) Rosaceae (rose) Verbenaceae (verbena) Sucrose ++++ ++++ +++ + ++++ ++ +++ +++ ++++ ++++ ++++ ++ +++ ++ Raffinose Stachyose Sugar alcohols + + Tr Tr Tr Tr Tr Tr ++ ++ +++ ++++ ++ Tr ++ + +++ Tr Tr Tr Tr + ++ ++ +++ Tr Tr ++++ + ++++ - • most families transport sucrose • concentration in phloem sap can reach 1 M Starch is made in photosynthetic and nonphotosynthetic cells Sel fotosintetik • cadangan sementara starch Triose P chloroplast •Pada daun hijau sucrose Sel non-fotosintetik • penyimpanan dalam jangka lama • akar, umbi, biji sucrose starch amyloplast Starch is the dominant storage polysaccharide in most plants Sunflower after 47 chloroplasts min photosynthesis In leaves - transitory starch - in high percentage of CO2 assimilated goes directly into starch Carbon absorbed (mg) 7.87 Hexose accumulated 1.17 Sucrose 4.20 Starch 1.84 • Pada sel non-fotosintetik- pati disimpan dalam amiloplas – storage organs – herbaceous – trees bananas, tubers (up to 80% dry weight), cereal grains (75% dry weight) roots, underground stems, bulbs perennials young twigs, roots, parenchyma of bark xylem & phloem Composition of Starch • Amilopektin – -1,4 & -1,6-glucan – 10,000 - 100,000 glucose units – highly branched, 20 - 25 glucoses/branch • Amilosa – -1,4-glucan – ~1000 glucose units acceptors for addition of further glucose units polymer of glucose units start (reducing end) • Butir pati – Water insoluble, – size & shape is species specific potato: oval, 100 µm in diameter rice: angular, 10 µm in diameter Fruktan • Beberapa tanaman menyimpan senyawa lain • Biasanya dalam bentuk fruktan – water-soluble, non reducing polymers of fructose – 5 - 300 fructose units, joined to one glucose • daun, bunga dan organ penyimpanan yg terletak dalam tanah – Asteraceae (dahlias, jerusalem artichokes) – Liliaceae (onions, asparagus) – Iridaceae (irises) • Daun pada beberapa Gramineae – C3 grasses - barley, oats, rye grass – major feedstuff for cattle & sheep in temperate zones – But store starch in the seed How are Sucrose and Starch Synthesised? Sukrosa dan pati disintesis pada tempat berbeda Pati dalam kloroplas Sukrosa dalam sitosol SINTESIS SUKROSA 1. Uridine-triphosphate (UTP) + Glucose-1-phosphate (G1P) ---> UDP-glucose + Pyrophosphate (PPi) 2. UDP-glucose + PPi + Fructose-6-phosphate (F6P) ---> UDP + sucrose-6(F)-phosphate (S6P) 3. Sucrose-6(F)-phosphate (S6P) + H2O ---> PPi + Sucrose SINTESIS UDP-Glukosa UTP CH2OH O OH Glucose 1-P PPi OH UDP-G pyrophosphorylase O O O-P-O-P-O-uridine OH O- O- PENGURAIAN SUKROSA Enzymes of Sucrose Metabolism Fructose 6P UDP-Glucose UDP Sucrose P Sucrose P Synthase Sucrose P Phosphatase Pi Sucrose Invertase Fructose + Glucose UDP Sucrose Synthase Fructose + UDP-Glucose polymer of glucose units start (reducing end) acceptors for addition of further glucose units BIOSINTESIS PATI PGI: phosphoglucose isomerase. PGM: Phosphoglucomutase; AGPase, ADPglucose pyrophosphorylase; SS, starch synthase; BE, branching enzyme; DBE, debranching enzyme; Glc-1-P, glucose-1-phosphate Enzymes of Starch Synthesis ATP ADPglucose Glucose 1-P PPi ADPG PPiase 1,4 glucann Glucose 1-P 1,4 glucann Starch synthase 1,4 glucann+1 Pi Starch phosphorylase Starch Branching Enzyme • Branching enzyme forms the -1,6 links start starch synthase start -1,4 link branching enzyme -1,6 link PENGURAIAN PATI Starch is catabolized by 5 enzymes: α-amylase, βamylase, α-glucosidase, starch phosphorylase, and αdextrin 6-glucanohydrolase (debranching enzyme) Genetic or biochemical modifications of starch are or may be used for... + amylose • fried snacks (crispness / browning) • thickener / gelling agent • biodegradable packing material • film coating Modified starch modified starch • Phosphate content • water absorbency • improve starch granule integrity (cross linker) + amylopectin • Improve freezethaw of frozen food • paper strength • adhesive • livestock feed addition