* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Biotechnology II PPT
DNA polymerase wikipedia , lookup
Metagenomics wikipedia , lookup
SNP genotyping wikipedia , lookup
Gene therapy wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Genealogical DNA test wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Non-coding DNA wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
DNA supercoil wikipedia , lookup
Epigenomics wikipedia , lookup
Genomic library wikipedia , lookup
Genome editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Designer baby wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic engineering wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Microevolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Molecular cloning wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
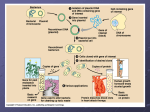
AP Biology Biotechnology Part 3 Bacterial Cloning Process Bacterium Gene inserted into plasmid Bacterial chromosome Cell containing gene of interest Plasmid Recombinant DNA (plasmid) Gene of interest Plasmid put into bacterial cell DNA of chromosome Recombinant bacterium Host cell grown in culture to form a clone of cells containing the “cloned” gene of interest Gene of interest Protein expressed by gene of interest Copies of gene Basic research on gene Gene for pest resistance inserted into plants Protein harvested Basic research and various applications Gene used to alter bacteria for cleaning up toxic waste Protein dissolves blood clots in heart attack therapy Basic research on protein Human growth hormone treats stunted growth Restriction Enzymes & Sticky ends Restriction enzymes are special enzymes, found in bacteria, that cut DNA at special places. The specific place that the DNA is cut is called a restriction site. When the restriction enzyme cuts the DNA at the restriction site, it creates fragments of DNA called restriction fragments. Restriction fragments have “sticky ends” that can match up with the ends of other fragments. Cut the DNA and Bacterial Plasmid The first step in bacterial cloning is to use specific restriction enzymes to cut the DNA and bacterial plasmid. This will cut both at the same place! Once the DNA has been cut, you can mix the fragments together and allow them to be joined using ligase. Reintroduce the Plasmid The next step is to reintroduce the recombined plasmid back into the bacteria. This is done through transformation bacteria picking up foreign DNA. The new bacteria are called a cloning vector. A vector is an organism that carries a recombined plasmid. Let the Bacteria grow and reproduce You will know let the bacteria grow and reproduce. The recombined bacteria will grow through binary fission and create new bacteria that should have the plasmid. Identify the transformed bacteria and culture the experimental bacteria Once you have grown the recombined bacteria, you will now need to isolate the experimental bacteria. 1. You will create a radioactive nucleic acid probe using phosphorous. 2. Then you will denature the DNA using heat to expose the bases. 3. The radioactive probe will join with the complimentary bases on the gene of interest. 4. Use a special film that will show the radioactive colonies and separate these from the others. Hershey-Chase Experiment Grow, verify protein production, and then use Source of the DNA for Transformation? Scientists must go from mRNA back to DNA to make the process easier. This is a tough thing to do because: - Prokaryotic DNA does not have introns. - Modified mRNA must be collected after it leaves the nucleus and turned back into DNA. - Use reverse transcriptase to turn single stranded RNA into double stranded DNA. You will need to first add a promotor sequence. - New cDNA (complimentary DNA) can be stored for use. INTRONS Yeast Artifical Chromosome (YAC)