* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Congenital Nystagmus
Genetic engineering wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Copy-number variation wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Neocentromere wikipedia , lookup
Public health genomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene therapy wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Human genome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene desert wikipedia , lookup
Population genetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Oncogenomics wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Medical genetics wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Helitron (biology) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
Genome editing wikipedia , lookup
X-inactivation wikipedia , lookup
Frameshift mutation wikipedia , lookup
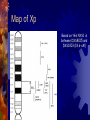
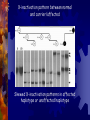
Congenital Nystagmus Erik Twaroski Eye gifs from <http://www.omlab.org> Characteristics Uncontrolled oscillation of the eyes Onset at birth or within several months 1 in 1,500 live births Patterns of inheritance autosomal dominant autosomal recessive X-linked dominant X-linked recessive X-linked dominant with incomplete penetrance Genes involved PAX6 NYS1 PAX6 Highly conserved sequence Positional cloning to determine area of genome (Hanson et al.) Maps to 11p13 Haploinsufficiency consistently present with all disorders associated with PAX6 All known mutations involve a single change in the amino acid sequence Mutation in both copies is lethal Thought to be the primary gene until recently PAX6 (cont.) Exons 4-13 contain coding regions. DNA binding domains and a linker. PAX6 mutations Sequence differences between normal and CN. Mutation creates a BsrI restriction site. Picture from Hanson et al. Mutation causing CN (Gly Val) shown in yellow at the N-terminal domain PAX6 (cont.) Complete loss of PAX6 in mice is lethal Knockouts cannot be made Elimination of one copy results is a small eye phenotype Picture from <http://www.mouse-genome.bcm.tmc.edu/ENU/publicimageview.asp> Another gene? Until 1999 PAX6 was believed to be the only gene responsible for CN All mutations resulted in eye disorders CN could only be linked to PAX6 NYS1 Cabot et al. first to report mapping CN to X chromosome Xp11.4-11.3 Dominant Important with incomplete penetrance for eye development Majority of research done on this gene Picture from Cabot et al. NYS1 (cont.) Started Xp by finding microsatellites on Sequences known from the Genome Database Regions of CA repeats Recombination events indicated which markers were closely linked Recombination events in parents of affected individuals Picture from Cabot et al. LOD scores for loci around NYS1 Picture from Cabot et al. More statistical analysis Support for location of an X-linked ICN gene, with respect to three chromosome Xp markers. Likelihood estimates are given in log10. Distances between marker loci, in centimorgans, are shown along the X-axis. The maximum location score for NYS1 is between DXS8015 and DXS1003, over the locus DXS993. Picture from Cabot et al. Map of Xp Based on this NYS1 is between DXS8015 and DXS1003 (18.6-cM) X-inactivation pattern between normal and carrier/affected Skewed X-inactivation patterns in affected haplotype or unaffected haplotype Treatments Currently no treatments available CN does not appear to interfere with visual function. Dell’Osso and Jacobs characterized the ocular oscillations of CN over a 35 year study published in July 2004 Treatments (cont.) Dell’Osso and Jacobs found that the body is able to compensate Even the most severe cases showed signs of some compensation More research needs to be conducted to further understand how the body is able to compensate Questions? References 1. Annick Cabet, Jean-Michel Rozet, Sylvie Gerber, Isabelle Perrault, Dominique Ducroq, Asmae Smahi, Eric Souied, Arnold Munnich, and Josseline Kaplan. “A Gene for X-Linked Idiopathic Congenital Nystagmus (NYS1) Maps to Chromosome Xp11.4-p11.3.” American Journal of Human Genetics 64:1141-1146, 1999. 2. Isabel Hanson, Amanda Churchill, James Love, Richard Axton, Tony Moore, Michael Clarke, Francoise Meire, and Veronica van Heyningen. “Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations.” Human Molecular Genetics, 1999, Vol. 8, No. 2. 3. Jonathan B. Jacobs and Louis F. Dell’Osso. “Congenital nystagmus: Hypotheses for its genesis and complex waveforms within a behavioral ocular motor system model.” Journal of Vision 4: 604-625, 2004. 4. Sanjaya Singh, Lian Y. Chao, Rajnikant Mishra, Jonathan Davies, and Grady F. Saunders. “Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function.” Human Molecular Genetics, 2001, Vol. 10, No. 9.