* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Rad51-deficient vertebrate cells accumulate
Epigenomics wikipedia , lookup
X-inactivation wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Molecular cloning wikipedia , lookup
Primary transcript wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Cancer epigenetics wikipedia , lookup
DNA vaccination wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genome editing wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Designer baby wikipedia , lookup
Helitron (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
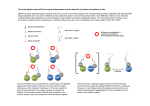
Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death BE.109 Module 1 Day 6 Manuscript discussion What is the normal function of the RAD51 gene? The RAD51 gene makes a protein also called RAD51, which is essential for the repair of damaged DNA. The protein made by the BRCA2 gene binds to and regulates the RAD51 protein to fix breaks in DNA. These breaks can be caused by natural or medical radiation. They also occur when chromosomes exchange genetic material (when pieces of chromosomes trade places) in preparation for cell division. The BRCA2 protein transports the RAD51 protein to sites of DNA damage in the cell nucleus. RAD51 then binds to the damaged DNA and encases it in a protein sheath, which is an essential first step in the repair process. In addition to its association with BRCA2, the RAD51 protein also interacts with the protein made by the BRCA1 gene. By repairing DNA, these three proteins play a role in maintaining the stability of the human genome. Useful reference: Tutt A and Ashworth A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol Med. 2002 Dec;8(12):571-6. [link] What is the normal function of the RAD51 gene? A model of the DNA-damage signal transduction cascade Tutt A and Ashworth A (2002) Trends Mol Med What is the normal function of the RAD51 gene? Tutt A and Ashworth A (2002) Trends Mol Med Where is the RAD51 gene located? The RAD51 gene is located on the long (q) arm of chromosome 15 at position 15.1. More precisely, the RAD51 gene is located from base pair 38,774,660 to base pair 38,811,645 on chromosome 15. What other names do people use for the RAD51 gene or gene products? • DNA repair protein RAD51 homolog 1 • HRAD51 • HsRAD51 • RAD51A • RAD51_HUMAN • RECA • RecA, E. coli, homolog of • RecA-like protein • recombination protein A What is the purpose of this study? To study functional role of Rad51 in the maintenance of chromosomal DNA during normal cell cycling. How can we study the function of Rad51? - By generating conditional RAD51 mutant clones tetracycline repressible promoter What is the experimental strategy? What are the neo and bsr resistance genes for? What is Southern blot? Why is it necessary for this study? More detailed explanation: http://www.accessexcellence.org/RC/VL/GG/southBlotg.html What is Western blot? Why is it necessary for this study? Useful link for blotting analysis: http://lifesciences.asu.edu/resources/mamajis/western/western.html What is the cell type the authors used? Why did they use it? The chicken B lymphocyte line DT40 - The high level of homologous recombination allowed us to perform targeted disruption of both RAD51 alleles. Cell cycle The cell cycle is an ordered set of events, culminating in cell growth and division into two daughter cells. Nondividing cells not considered to be in the cell cycle. The stages, pictured to the left, are G1-S-G2-M. The G1 stage stands for "GAP 1". The S stage stands for "Synthesis". This is the stage when DNA replication occurs. The G2 stage stands for "GAP 2". The M stage stands for "mitosis", and is when nuclear (chromosomes separate) and cytoplasmic (cytokinesis) division occur. Image: http://www.bmb.psu.edu/courses/biotc489/notes/ How can we analyze the cell cycle? A common way of ascertaining a proportion of a population of cells at any given part of the cell cycle is to analyse according to the DNA content (by definition G2 cells have twice as much DNA as G1). The process requires cell permeabilisation, RNAse treatment and staining (most commonly with PI however other stains such as DAPI and 7AAD are also used) prior to analysis. Following exclusion of cell doublets and clumps from the analysis, a healthy, growing cell population will exhibit a characteristic histogram profile as above. The analysis of this histogram by the pragmatic approach will then ascertain the proportional cell numbers in G1-S-G2/M phases and can also be used to measure nuclear condensation in apoptotic cells. Bromodeoxyuridine (BrdU) Incorporation BrdU is an analogue of thymidine and will be taken up into the DNA of cycling cells. To detect this, we can unwind the DNA (by using acid, alkali or enzyme) and then use an antibody against BrdU. In this way, we can separate G1, S and G2 cells. Obviously BrdUpositive cells will equate to S phase cells only when the labeling time is short as cells in late S at the beginning of the labeling period. Bromodeoxyuridine (BrdU) Incorporation Control (left) versus drug treated (right) effect on cell cycle. How is replication related with homologous recombination? • Animation file made by Justin H. Lo may help you understand how replication is related with homologous recombination: http://web.mit.edu/jhlo/www/urop/Fork_Col lapse_a.swf Isochromatid-type gaps and breaks Both sister chromatids of a single chromosome are broken at the same locus Useful link: http://www.inchem.org/documents/ehc/ehc/ehc46.htm