* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download blumberg-lab.bio.uci.edu
Biology and consumer behaviour wikipedia , lookup
Primary transcript wikipedia , lookup
Microevolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Protein moonlighting wikipedia , lookup
Genome (book) wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Epigenetics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Neocentromere wikipedia , lookup
Genome evolution wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Minimal genome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Epigenetics of human development wikipedia , lookup
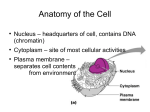
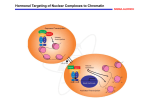
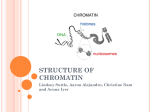
Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells Guillaume J. Filion, Joke G. van Bemmel, Ulrich Braunschweig, Wendy Talhout, Jop Kind, Lucas D. Ward, Wim Brugman, Ines J. de Castro, Ron M. Kerkhoven, Harmen J. Bussemaker, and Bas van Steensel Dristi Angdembey Anton Arce D145 Chromatin: structure and general definition http://www.nature.com/scitable/content/ne0000/ne0000/ne0000/ne0000/113158606/18847_6.jpg Traditional classification 2 types of chromatin: 1. Heterochromatin o repressed genes o dark staining = tighter packing 2. Euchromatin o An electron micrograph of a nucleus http://www.histology.leeds.ac.uk/cell/nucleus.php expressed genes Distinct types of heterochromatin discovered in Drosophila 1. PcG chromatin o o Polycomb group (PcG) proteins + methylation of lysine 27 of histone H3 (H3K27) repressive type that regulates genes with developmental functions ● Sparmann and van Lohuizen, 2006 2. HP1 chromatin Heterochromatin protein 1 (HP1) + associated proteins + methylation of H3K9 o found around the centromere o reporter genes integrated in or near likely repressed o ● Hediger and Gasser, 2006 Properties of distinct chromatin types 2 important principles of chromatin organization: 1. domains with distinct protein compositions 2. coverage of long stretches of DNA Are there other types of chromatin that follow these principles? Genome-wide location maps of 53 Chromatin Proteins ● Embryonic D. melanogaster cell line Kc167 used ● Representative cross-section of chromatin proteome ● Used new technique DNA adenine methyltransferase Identification http://www.entu.cas.cz/ , https://www.broadinstitute.org/ , http://www.wikimedia.org/wikipedia/commons DNA adenine methyltransferase Identification (DamID) ● Fusion protein of E. coli Dam and target chromatin protein ● Methylates adenines of nearby GATC sequences ● Uses microarray to ID http://research.nki.nl/Vansteensellab/DamID_FAQ.htm methylated A’s ● Sensitive to transient DNA interactions ● Has 1 kbp resolution ● Goes over whole genome 300 bp at a time Why not ChIP? ● Not great for large-scale application o paper probes for 53 proteins ● Dependent on quality and specificity of antibodies ● Dependent on crosslinking reagents o inability to bind to chromatin proteins with short residence times http://www.usbweb.com/assets/78460_fig1.gif Chromatin Binding Map Chromatin Types by Arbitrary Color ● GREEN: HP 1 chromatin; classic heterochromatin corresponds to the heterochromatin ● BLUE: PcG chromatin discussed earlier ● BLACK: Half of genome, very repressive, related to nuclear lamina, late replicating ● RED: Euchromatin-like, “hub” of cell regulation, DBF hot spot, replication origins ● YELLOW: Euchromatin-like, constitutive “housekeeping” genes, DNA repair ‘GREEN’ Chromatin ● ● ● ● corresponds to classic heterochromatin, i.e. HP1 chromatin marked by SU(VAR)3-9, HP1, and HP-1 interacting proteins prominent in pericentric regions and on chromosone 4 H3K9me2, histone mark generated by SU(VAR)3-9, highly and specifically enriched in GREEN chromatin ‘BLUE’ Chromatin ● corresponds to PcG chromatin o extensive binding by the PcG proteins: PC, E(Z), PCL, SCE ● well-known PcG target loci such as the Hox gene clusters are localized in BLUE domains ● H3K27me3, histone mark generated by E(Z), highly enriched in BLUE chromatin ‘BLACK’ Chromatin ● ● ● ● The most prevalent type of chromatin Least genes per genome coverage Most silent genes, with no mRNA for 66% Genes are actively repressed How did they figure out BLACK Chromatin is repressive? The question: “Does BLACK chromatin actively repress transcription or merely forms secondary to a lack of transcription?” ● if repressive, transgenes inserted into BLACK chromatin may exhibit reduced transcription ● if secondary, transgenes should be unaffected Developmental Regulation of Genes in BLACK chromatin ● 33% of non-silent genes in BLACK chromatin o tissue-specific expression ● suggests that BLACK chromatin domains can be remodeled into a different chromatin type in some cell types ● rich in Highly Conserved Noncoding Elements (HCNEs) o mediate gene regulation? ‘RED’ and ‘YELLOW’ Chromatin ● Most like classic euchromatin o o o High rates of transcription and polymerase activity High gene density Replicated first ● Share a set of proteins that make them like euchromatin ○ ○ ○ Histone Deacetylases: remove acetylene, regulates expression ASH2: Methyltransferase, regulates expression DF31: Chromatin decondensation ‘RED’ Chromatin ● Is replicated first and has most origins of replication ● Concentrates nucleosome remodeling ● Contains key protein in chromosome structure ● Genes are linked to behavior and tissue specific processes ‘YELLOW’ Chromatin ● Lots of histone H3K36me3 with chromodomain modification o Leads to constitutive activity ● Genes transcribed are widely expressed and include universal cell processes Predicting DNA binding from Chromatin Types ● Genome has more binding motifs than DBFs actually bind to ● Guided by chromatin type via interacting chromatin proteins ● Specific to organism as chromatin does not interact with foreign protein Future directions ● How do these principal chromatin types differ between cell types and species? o Do species with more “complex” genome have more chromatin types? Drosophila: 4 chromosomes Humans: 23 chromosomes ● Do the chromatin types contribute to epigenetic memory? Mechanism? DamID- shortfalls/disadvantages ● Not standardized, mainly uses microarrays not Nextgen ● Not perfect targeting leads to background artifacts o Non-specific binding in more “open” chromatin or high concentration of GATC ● High Dam concentrations leads to overmethylation o Needs weak induction because of high activity Take this home guys ● DamID is a good option for large chromatin protein sets with lots of coverage ● Chromatin is more diverse than euchromatin and heterochromatin o o Unique protein content leads to different expression and repression of genes Protein DNA binding is dependent on chromatin type Acknowledgement ● Professor Blumberg ● Bassem Shoucri