* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Artificial gene synthesis wikipedia , lookup
Genomic imprinting wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Neocentromere wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding DNA wikipedia , lookup
Oncogenomics wikipedia , lookup
X-inactivation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Human genetic variation wikipedia , lookup
Microsatellite wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Frameshift mutation wikipedia , lookup
Koinophilia wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genetic drift wikipedia , lookup
Point mutation wikipedia , lookup
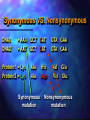
Dominance • Refers to an allele’s phenotypic effect in the heterozygous condition • NOT its numerical prevalence • For example, a Dominant allele may be LESS common than a recessive allele! Dominance • If the dominant and recessive alleles A and a (with freq. of p and q) are in H-W Equilibrium • The frequency of the dominant phenotype is p2 and 2pq • The frequency of the recessive phenotype is q2 Dominance – Moth Example • Black Phenotype (Dominant) = 10% • Grey Phenotype (Recessive) = 90% p=Black allele q=Grey allele • Assume this population H-W Equil. (Rounding) • q2 = 0.9 q = 0.95 (0.94868) • therefore p = 0.05 (0.051316) • This implies that p2 = (0.05)2 = 0.0025 (0.0026334) • .25% are dominant homozygote • Heterozygotes = 2pq = 2(0.05)(0.95) = 0.095 0.095 (or 9.5%) are heterozygotes Dominance • A dominant allele may well be less common than a recessive allele. Estimating the Proportion of Polymorphic Loci • Why? To know how much genetic variation exists within a population -Also, allows us to understand patterns of variation (as we will see in a few minutes...) • HOW? Methods Used to Estimate Genetic Polymorphism • Protein electrophoresis • RFLP’s restriction fragment length polymorphisms • DNA Sequences • Microsatellites Polymorphism of Adh Polymorphism of Ldh Variation at the DNA Level Allelic diversity across loci • Heterozygosity or mean heterozygosity • As the avg. frequency of heterozygotes across loci • As the fraction of loci that are heterozygous in the genotype of the avg. individual. • % of polymorphic loci • Fraction of loci in a population that have multiple alleles. Heterozygosity Genetic variation at allozyme loci How is polymorphism maintained? • Do forces of natural selection maintain these polymorphisms, or are they neutral, subject only to the operation of random genetic drift? Multiple Loci •Genes do not exist in isolation •Multiple genes probably effect a single character •Physically associated on chromosome •Linkage disequilibrium Evolution at Multiple Loci • We have considered the Hardy-Weinberg • • Equilibrium Principle for one locus at a time Can this model be extended to a two or higher locus case? Must make a more realistic model Evolution at Two Loci • Consider two loci on the same chromosome: A and B • Alleles A and a; B and b • Track both allele frequencies and • chromosome frequencies Four possible chromosome genotypes: AB, Ab, aB, and ab • Haplotypes: multilocus genotype of a chromosome Evolution at Two Loci • Loci are in linkage equilibrium if the • frequency of one allele does not affect the frequency of the other Linkage disequilibrium = locus frequencies affect each other • May affect evolution of each other by genetic linkage • Genes may be inherited as a group • May be based on physical distance between the loci Evolution at Two Loci • Numerical example • Two hypothetical populations with 25 • chromosomes each We can calculate allele frequencies and chromosome frequencies Evolution at Two Loci • Numerical example • Allele frequencies same for both populations • Chromosome frequencies differ slightly • To see difference, can calculate frequency of allele B on chromosomes carrying A versus frequency of chromosomes carrying a • Can depict this graphically Evolution at Two Loci • Numerical example • Top population is in linkage equilibrium • Chromosome genotype of one locus is independent of other locus • Bottom population is in linkage disequilibrium • Nonrandom association between a genotype at one locus and another • If we know one genotype we have a clue about the other Evolution at Two Loci • Conditions for Linkage Equilibrium • The frequency of B on chromosomes • • carrying A is equal to the frequency of B on chromosomes carrying a The frequency of any chromosome haplotype can be calculated by multiplying frequencies of constituent alleles The quantity D, the coefficient of linkage disequilibrium, is equal to zero • D = gABgab - gAbgaB • gs are frequencies of chromosomes Evolution at Two Loci • Assess conditions for hypothetical example • Frequencies of chromosomes are equal • True for top, not for bottom • Frequencies of haplotypes can be calculated by multiplying allele frequencies • A X B = (0.6)(0.8) = 0.48 • A X B = (0.6)(0.8) = 0.48 • D=0 YES NO AB = 0.44 • gABgab - gAbgaB = (0.48)(0.08) = (0.12)(0.32) • gABgab - gAbgaB = (0.44)(0.04) = (0.16)(0.36) YES NO Evolution at Two Loci • If the population is in linkage equilibrium, Hardy-Weinberg equations can be used for each locus independently • Assume no selection, no mutation, no migration, infinite population, panmixia • What creates linkage disequilibrium? • Selection on multilocus genotypes • Genetic drift • Population admixture Evolution at Two Loci • How to eliminate linkage disequilibrium • Sexual reproduction reduces linkage • • • disequilibrium Meiosis, crossing over, outbreeding Meiosis breaks up old genotype combinations and creates new ones Genetic Recombination • Randomizes genotypes of loci with respect to each other Evolution at Two Loci • Why does linkage disequilibrium matter? • If two loci are in linkage disequilibrium, • • selection at one locus changes allele frequencies at the other Cannot use Hardy-Weinberg to calculate allele or genotype frequencies In practice the change in one locus due to linkage disequilibrium could erroneously be interpreted as selection on that locus Evolution at Two Loci • Why does linkage disequilibrium matter? • If in linkage equilibrium Hardy-Weinberg can • still be used Random sexual reproduction is very efficient at eliminating linkage disequilibrium • Most loci are in linkage equilibrium • Empirical study of 5000 human loci found that only 4% were in linkage disequilibrium Evolution at Two Loci • CCR5-D32 allele • • • • • • Where did the allele come from? Why is it mainly in Europe? Stephens measured linkage disequilibrium in CCR5D32 with two loci nearby on same chromosome • GAAT and AFMB • Neutral alleles 192 Europeans GAAT and AFMB are nearly in linkage equilibrium CCR5-D32 in strong linkage disequilibrium with both Evolution at Two Loci • CCR5-D32 allele • Linked alleles • + - 197 - 215 • How did linkage disequilibrium arise? • Selection, genetic drift, or population admixture • Not selection because GAAT and AFMB are • • selectively neutral Not population admixture or the allele would be elsewhere in the world Must be genetic drift Evolution at Two Loci • CCR5-D32 allele • At some time in past only the wild type allele • • (+) existed Then on the chromosome CCR5-GAAT-AFMB with alleles + - 197 - 215 a mutation occurred to D32 Linkage is now breaking down • Some individuals with + - 197 - 217 are now found Evolution at Two Loci • CCR5-D32 allele • Stephens used rates of crossing over and • • mutation to calculate how fast the linkage disequilibrium would be expected to break down Used this calculation to estimate how long ago D32 appeared This allele first appeared between 275 and 1875 years ago • Probably about 700 years ago Evolution at Two Loci • CCR5-D32 allele • Unique mutation must have happened in • • • Europe Probably occurred elsewhere but was not favored by selection Why was D32 favored in Europe? Must have been strong selection to raise from nearly 0% to 20% in 700 years Evolution at Two Loci • CCR5-D32 allele • Perhaps D32 provides protection from other • • diseases It has been hypothesized that it protects against the bacterium Yersinia pestis, the pathogen that caused the Black Death Investigations are being performed now to test this theory Geographic Variation • Differences in phenotype (or genotype) among different geographic populations of the same species Patterns of Geographic Variation • Sympatric Populations • (syn = “together”; patra = “fatherland”) • Overlapping ranges • Parapatric Populations • (para = “besides”) • adjacent but not overlapping ranges • e.g. High elevation and low elevation separate populations • Allopatric Populations • (allos = “other”) • non-overlapping Rat Snakes (Elaphe obsoleta) Allopatric geographic variation Molecular Phylogeny of Rat Snakes (Elaphe obsoleta) Historical dispersal patterns in Rat Snakes (Elaphe obsoleta) Genetic Distance • Quantify the degree of genetic differentiation among two or more populations of the same species, or among different species. • Nei’s D Geographically Variable Characters • Morphological • Life history • Behavior • Ecology Character Displacement • Sympatric populations of two species differ more than allopatric populations in their food use etc. these differences are generally reflected in morphology. Conclusions about variation • A species is not genetically uniform over its geographic range • Some differences appear to be adaptive consequences • Genetic differences between populations are the same in kind as genetic differences among individuals within a pop. Origin of Genetic Variation • Gene mutation – an alteration of the genetic (DNA) sequence • Mutations have evolutionary consequences only if they are transmitted to the next generation • Somatic cell • Germ line cell Various types of mutations • Point mutations • Transition – substitution of a purine for a purine or a pyrimidine for pyrimidine (A G) or (C T) • Transversion – substitution of a purine for a pyrimidine or vice-versa (A T) or (A C) ... and so-on Categories of Point Mutations • Synonymous • Change in DNA sequence No Change in Amino Acid sequence • Non-Synonymous • Change in DNA sequence Change in Amino Acid sequence Synonymous VS. Nonsynonymous DNA1 DNA2 = AAA GCT CAT = AAT GCT GAT Protein1 = Lys Protein2 = Lys Ala Ala Synonymous mutation His Asp GTA GAA GTA GAA Val Val Glu Glu Nonsynonymous mutation Various types of mutations • Frameshift Mutations • Where nucleotides in the DNA sequence are added or deleted, creating a change in the reading frame for the protein encoded by the gene Frameshift Mutations Transposable elements • Sequences that can move to any of the many places in the genome • Barbara McClintock Maize Transposable elements • Carry genes that code for transposases. • Carryout transposition • Conservative transposition • Replicative transposition • Process of insertion generates 4-12bp repeats of the host DNA at the end of the element. Transposable elements • Insertion sequences: 700-2600bp only code for transposases • Transposons: 500-7000bp encode transposases + other functional genes. • Retroelements: use reverse transcriptase to transcribe. HIV like. Except they do not cross cell boundaries and spread only by cell division Effects on host genome • Increase total genome size • Alteration of host gene expression • Increase in mutation rate of host genes • Chromosome rearrangements (host) • Retrogenes. Reverse transcription • Most non functional • psuedogenes Mutation Rates • Estimates of u (mutation) depend on how you calculate them • Phenotypic studies • White vs red eye in Drosophila • DNA sequence studies • Count number between 2 closely related organisms Mutation estimates • Usually count the number of mutations, scored my their phenotypic effects among the offspring of an initially homozygous stock. • In Drosophila and other multi-cell organisms usually measure # of chromosomes bearing a particular mutation from a homozygous population for an allele. Use special crosses to visualize phenotypic effect. Mutation estimates • The autosomal chromosomes of N flies represents 2N gametes that form them, so mutation rate is calculated a number of new mutations per gamete per generation • Avg. locus mutation rate based on phenotypic effects 10-6 - 10-5 mutations per gamete per generation Mutation Rates • Back mutations-mutation of an allele from the mutant type back to “wild type” • Multiple hits; multiple substitutions at same sit. Estimating Substitution rate from DNA sequence Outgroup) Taxon 1) Taxon 2) Taxon 3) Taxon 4) ATGTCAGGGACTCAGATCGAATGGGATCTAG .....C......T.................. .....G......T........C......... .....C...........A............. .....G...........A........G.... Average mutation rate per base pair has been estimated at 10-9 A G C Purines T Pyrimidines Substitutions Time 0 Outgroup) Taxon 1) Taxon 2) Taxon 3) Taxon 4) ATGTCAGGGACTCAGATCGAATGGGATCTAG .....C......T.................. .....G......T........C......... .....C...........A............. .....G...........A........G.... Substitutions Time 1 Outgroup) Taxon 1) Taxon 2) Taxon 3) Taxon 4) ATGTCAGGGACTCAGATCGAATGGGATCTAG .....A......T.................. .....G......G........C......... .....G...........A............. .....G...........A........G.... Substitutions Time 2 Outgroup) Taxon 1) Taxon 2) Taxon 3) Taxon 4) ATGTCAGGGACTCAGATCGAATGGGATCTAG .....G......T.................. .....G......T........C......... .....G...........A............. .....G...........A........G.... Multiple Substitutions at the same site Evolutionary implications of Mutation rates • Avg. per-locus mutation rate of 10-5 (1 in 100,000 generations) is so low that the rate of change in the frequency of an allele, due to mutation alone, is very low. • Phenotypically distinguishable alleles A1 and A2 Evolutionary implications of Mutation rates • These have allele freq. p(=1-q) and q. • A1 mutates to A2 at rate u= 10-5 • Therefore in each generation, the frequency of A2 is increased by u x p (that is, a fraction u of A1 alleles mutate. • Thus change in freq of A2 ∆q = up=u(1-q) Mutation rates • q = 0.5 • In the following generation • q’ = q + ∆q = • ∆q = up = 10-5 (0.5) = 0.000005 • 0.5 + 0.000005 = 0.5000005 Mutation rates • How does mutation affect the freq. of alleles on a pop. • assume no back mutation e-ut pt = po qt = 1-poe-ut u = forward mutation t = # of generations q = .5 change q = .75 = 70,000 generations q = .75 change q = .875 = 70,000 generations Mutation rates • Very low mutation rate • Therefore mutation alone will probably not account for the actual speed of evolutionary change observed in nature. Mutation rates • But 150,000 genes in humans • 10-5 mutations per gene x 105 genes = 1 mutation per haploid genome in humans. • Probably an underestimate • If even a tiny fraction of there were advantageous, the amount of new “raw material” is substantial. Mutation rates • Neutral Theory of molecular evolution • Mutations that neither enhance nor lower fitness • Fixed (go to 1) or lost (go to 0) entirely by chance • The probability that this will occur is u • That is, a mutation that occurred in the past will become fixed • After the passage of t generations the mutations that have become fixed would equal x = ut Mutation rates • If 2 species diverged from a common ancestor t generations ago, the expected fraction of fixed mutations is D = 2ut • If we are talking bps then a fraction of D = 2ut of the bps of a gene should differ between species, assuming equal prob. of mutation. Mutation rates • Avg. mutation rate per bp per generation is u = D/2t • Where D is the number of bp differences between 2 sequences • Human chimp split 1.3 x 10-9 per site per year (7mill) assuming 15-20 years generat. Mutations • Then the mutation rate is 2 x10 -8 per generation. • Human diploid genome has 6 x 109 nucleotide pairs, so this implies 120 new mutations per genome per generation. • Page 273 Futuyma