* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Hedgehog signaling pathway wikipedia , lookup
Signal transduction wikipedia , lookup
P-type ATPase wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Protein domain wikipedia , lookup
Promoter (genetics) wikipedia , lookup
VLDL receptor wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Transcription factor wikipedia , lookup
Gene regulatory network wikipedia , lookup
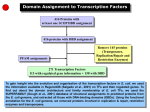
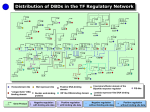
BACKGROUND E. coli is a free living, gram negative bacterium which colonizes the lower gut of animals. Since it is a model organism, a lot of experimental data is available. Furthermore, the complete genome sequence (Blattner et al., 1997) of this microbe has enormously helped to integrate a vast amount of biological information. In this work we analyse an important class of molecules namely transcription factors which regulate gene expression. We study their domain architecture to understand their evolution, their regulatory function as transcriptional activators or repressors and the evolution of the regulatory network. DOMAINS are the structural and evolutionary unit of proteins as defined in the SCOP database (Murzin et al., 1995) and a FAMILY is a set of related domains which originated from a common ancestor. OBJECTIVES What are the different protein families that constitute the E. coli transcription factors? Are regulatory functions related to domain architecture or binding site position? How complex is the gene regulatory network in E. coli and how did it evolve? RESULTS There are 11 different DNA-binding domain (DBD) families and 46 different partner domains in 271 transcription factors (TFs) On average each TF has 2 domains: one DBD and a partner domain which is generally a control domain 73% of the TFs have arisen by gene duplication Position of the TF binding site is the determining factor for activation or repression rather than the DBD type, partner domain or domain architecture Transcription factors vary in the number of genes they regulate. The dominant transcription factors that regulate the largest number of genes also regulate the largest number of TFs to amplify their influence 1/3 of the interactions have homologous transcription factors that share a set of regulated genes or regulated genes that share a set of transcription factors. This suggests that duplication is a major mechanism in the growth of the gene regulatory network Domain Architectures of E. coli TFs 271 TFs have 74 distinct domain architecture 73% of E. coli TFs have arisen by gene duplication Activators, Repressors and Dual Regulators Domain architecture, DNA binding domain or partner domain type is NOT indicative of the regulatory function Distance of the TF binding site from the transcription start site is the determining factor 33% of repressor binding sites occur after the transcription start site