* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Gene regulation_1130(final)
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
History of genetic engineering wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Primary transcript wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Microevolution wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
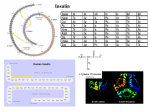
Gene regulation 1 Insulin • Insulin affects the expression of many more genes (>200) • Insulin affects transcription, mRNA stability, and translation (from gene to protein) • Focusing on insulin-regulated gene transcription – Positive and negative effects on the transcription of specific genes even within the same cell. – Insulin-regulated genes coding proteins involved in a variety of biologic phenomena (F25.1) – liver, muscle, adipose tissue >> other tissue 2 F 25.1 Insulin regulated genes 3 Cis/trans model for insulin regulated gene transcription • Transcription is mediated by the interaction of cis-acting DNA elements with trans-acting factors. • Specific sequence of the cis-acting element determines which trans-acting factor will bind. => These cis-acting elements are referred to as insulin response sequences or elements (IRSs/IREs) 4 5 • Impaired insulin-regulated gene expression decreased : peripheral glucose utilization : rate of insulin secretion 6 Specific defects in gene expression in the etiology of in MODY • • • • MODY-1: Hepatocyte nuclear factor (HNF)-4 MODY-3: HNF-1α MODY-4: IPF-1 MODY-5: HNF-1β => Complex : these genes encodes a transcription factor that could regulate the expression of multiple islet genes. • MODY-2: glucokinase => a key element in glucose sensing by the Betacell 7 How a signal passes from the insulin receptor in the plasma membrane to a specific trans-acting factor binding to an IRS ? * Insulin actions on gene transcription by two ways A) through plasma membrane-initiated changes in nuclear protein phosphorylation /dephosphorylation B) more directly via intracellular insulin receptors. • Recent reports suggest – cell surface receptors translocate to the nucleus. – Insulin receptor substrate-1 and 3 in the nucleus. – PI3-kinase/PKB signaling system in the nucleus. 8 Identification of positive insulin response sequences 1. 2. 3. 4. 5. The serum response element The Ets motif The Activator protein-1 motif The Sp1 Motif The Sterol response element and the E-Box motif 6. The Thyroid Transcription Factor-2 Motif 7. Miscellaneous 9 The serum response element • C-fos SRE – C-fos gene is cellular homologue of the transforming gene of FBJ murine osteosarcoma virus – Insulin stimulates c-fos gene transcription in cell system – -356~+109 of c-fos promoter sequence mediates the insulin response • SRES in the β-actin and GLUT-1 – Mediate positive effects of insulin => SRE mediates effects of several hormones on gene transcription rather than insulin specific effect 10 Signal transmission from insulin receptor in plasma membrane to c-fos IRS in nucleus • Insulin Insulin Rc. in plasma membrane Shc (thyrosinephosphorylated IRS family member) binds GRB-2 (adaptor molecule) mSOS (guanine nucleotide exchange factor) p21ras 형성을 촉진 raf-1 (serine /threonine kinase) 활성화 MEK1/2 활성화 Erk1/2 p90RSK cFOS (p211 참조) RSK cFos • Thus, ERK1/2 kinase pathway involves in insulin-stimulated c-fos gene transcription – MEK1/2 inhibitor (PD98059) blocks insulin-stimulated c-fos expression => However, target of insulin signaling at the c-fos remains unclear 11 SRE binding proteins • • p67SRF, C/EBP-β, Phox 1, DBF/MAPF1 ERK1/2 kinase pathway 는 p62TCF 를 target으로 하며, 인슐린에 의해 활성화 된 다른 경로는 p67SRF을 Target 으로 한다. => insulin can regulate c-fos gene transcription either directly through the SRE or through the 5’ Ets motif. 5’ Ets motif p62TCF Erk1/2 C-fos SRE p67SRF Other pathway 12 The Ets Motif • 인슐린에 의한 gene expression 이 Ets motif를 통해서 일 어남은 prolactin gene 에서 처음 보고되었다 (in the GH3 rat pituitary cell line) – Prolactin IRS was mapped to a region of the rat promoter between -212~+73 – Primary insulin response sequence: -97~-87 – For the full insulin response: -76~-67 also required • Prolactin IRS is composed of the dyad repeat sequence CGGAA(A/G)(T/A)(A/G)CAT with a 10-bp intervening sequence. – CGGAA is critical sequence • Ets motif is important for insulin-induced somatostatin and thymidine kinase gene transcription 13 • Prolactin IRS에 binding 하는 Ets family : GABP-α, GABP-β1, Ets-1, Elk-1, CREbinding protein (CREB) • Pit-1 (pituitary specific factor) – when Pit-1 binding sites are mutated, the stimulatory effect of insulin on prolactin gene transcription is reduced – Deletion of Pit-1 binding sites dramatically reduces basal prolactin gene transcription – accessory factor vs. a direct target of insulin signaling 14 PI 3-kinase pathway: most important for metabolic action of insulin • PI 3-kinase pathway가 Gene transcription 을 포함한 insulin action에 있어서 가장 중요한 것으로 알려져 있다. • PI-3kinase는 110kd catalytic subunit와 85-kd의 regulatory subunit로 구성되어 있으며, p85 subunit의 두 개의 SH2 domain 이 IRS-1과 상호 작용을 하고, 결과적 으로 p110 을 활성화 시킴. • Wortmannin, a specific inhibitor of PI 3-kinase, blocks many of insulin’s actions in various cell types. => PI 3-kinase is required for the insulin mediated prolactin gene transcription. 16 The Activator Protein-1 Motif • AP-1 motifs are important for the gene expression by insulin; 1. Matrix metalloproteinases (MMPs). – First documented – Accessory factor가 필요: Ets motif 2. malic enzyme 3. hepatitis B virus X gene 17 Action of insulin through the AP-1 motif at two levels (F25.2) => Insulin has both short- and long-term effects on gene expression through the same element. 18 • AP-1을 매개로 한 인슐린의 효과는 ERK1/2 kinase pathway를 필요로 한다 • But it depends on cell type – In CHO. T cell: ERK1/2 kinase activation is sufficient – In adipocytes: FRAP/mTOR pathway activation is also required • 최근에 Maf 와 activating transcription factor (ATF) 도 AP-1 motif에 결합하여 작 용을 나타냄을 보고. – target gene은 아직 밝혀지지 않음 19 The Sp1 Motif • Insulin-regulated calmodulin gene transcription 을 통해 처음 밝혀짐 • Delta1-crystallin, PAI-1, apolipoprotein A1 are stimulated by insulin through Sp1 • Interesting issues regarding Sp1 1. Sp1 binds the promoters of many genes, (not all) which are regulated by insulin. 2. Sp1 may oppose the action of insulin 3. Sp1 can also mediate effects of glucose on gene expression. 20 • Sp1 belong to highly related family members: Sp2, Sp3, Sp4 (may be much larger) • Mechanism of Sp1 in insulin action: complex – Sp1 is phosphorylated by multiple protein kinases, known to be insulin regulated. – Insulin can also regulate Sp1 gene expression. • Insulin induces Egr-1 gene expression – Sp1과 Egr-1은 비슷한 DNA sequence를 인지 함으로 DNA binding 에 있어서 경쟁을 할 수 있고 이러한 점 때문에 인슐린이 복잡한 효과를 나타냄. – 예) malic enzyme promoter에서 Sp1은 인슐린 signaling 을 촉진 하지만, Egr-1은 이를 억제함 • 또한 Sp1 은 인슐린에 의해 조절되는 GAPDH gene의 발현에 관여하는 것으로 보고됨 21 The Sterol Response Element and the E-box Motif • Unusual transcription factor – SREBPs can bind two motifs: sterol response element and the E-box motif. • Released from the Endoplasmic reticulum • SREBP-1a, SREBP-1c, SREBP-2 – SREBP-1 activates genes required for fatty acid synthesis – SREBP-2 activates genes required for cholesterol synthesis • Insulin preferentially induces SREBP-1c, but , through the stimulation of the ERK1/2 kinase pathway, might activate both SREBP-1 and SREBP-2 22 • Insulin은 SREBP-1c 를 통하여 직접적으로 fatty acid biosynthesis 에 관여하는 multiple hepatic genes 의 발현 에 관여한다고 알려져 있지만, 실제적으로 glucose 와 insulin 이 둘 다 높을 때 이러한 gene들이 발현함. => Glucose rather than insulin was the key signaling molecule in hepatic gene expression • Kahn demonstrated that glucose was sufficient for the induction of pyruvate kinase gene expression in primary hepatocytes. => insulin mainly play a permissive role in the regulation of hepatic gene expression through – stimulating glucokinase gene transcription – promoting glucose phosphorylation. 23 fatty acid synthase (FAS) • FAS: central role in de novo lipogenesis in mammal. • Insulin increases FAS gene transcription in the livers of diabetic mice. • IRS of FAS is located in a region between -332 and +67 in the FAS promoter. • E-box motif mediates the effect of insulin through Fas. 25 Relative roles of glucose and insulin in the regulation of FAS gene expression 1. Insulin/glucose → through the proximal region of the FAS promoter → 2 to 3-fold stimulation of FAS gene expression • fasting/refeeding → 20-30 fold induction of the endogenous FAS gene => unidentified upstream FAS promoter region must be important for the fasting/refeeding response. 2. SREBP-1c is not a direct target of glucose signaling itself and other transcription factors mediate the action of glucose on FAS gene expression. • Full induction of FAS gene expression requires 1) glucose action, mediated through a carbohydrate-responsive factor 2) insulin action, mediated through SREBP-1c 26 Upstream stimulatory factor (USF) • Insulin-regulated FAS gene transcription involves another transcription factor, USF • Sul demonstrated that E-box motif (-68 to -52 FAS promoter region) mediates the effect of insulin (in tissue culture cells) – Selective mutations in the SRE (E-box intact) had no effect on the insulin response – Instead, the binding of USF1/2 to E-box correlated with the effect of insulin on FAS gene transcription (in 3T3-L1 adipocytes) 27 • In contrast, Osborne demonstrated that mutation of the SRE (intact E-box motif) blunted insulin-stimulated FAS gene expression => Conflicting data ! • Possible solution => SREBPs have a dual binding specificity for both SREs and E-box motifs in their DNA binding domains . 28 The Thyroid Transcription Factor-2 Motif (TTF-2) • Thyroglobulin (Tg), the macromolecular precursor of thyroid hormone, is the major protein synthesized by thyroid cells. • IGF-1 과 insulin 은 이러한 Tg gene transcription 을 자극 하며, 이러한 효과는 Tg promoter 의 -168~+39 region에 의해 매개 • 이 region 내에 3개의 transcription factor binding sites가 존재: TTF-1, TTF-2, ubiquitous factor (UFA) • Insulin & IGF-1: dramatically stimulate binding of TTF-2 • TTF-2 gene transcription 은 thyroperoxidase expression (enzyme involved in the synthesis of thyroid hormone) 에 도 필요 (TTF-2만으로는 부족하고, TTF-1과 ubiquitous factor (UFB) 도 있어야 한다). 29 UFB (nuclear factor-1) 1. Insulin also stimulates NF-1 expression in thyroid cells => indicating that insulin regulates thyroperoxidase gene expression through a combination of two transcription factors, TTF-2 and NF-1. 2. Insulin also stimulates expression of an NF-1 in liver cells. 3. 그런데, NF-1 이 지방세포에서 인슐린의 GLUT-4 의 발현 에 대한 negative effect를 매개 * NF-1 의 역할은 아직 명확하지 않아 IRS로 구분하지는 않 음 30 Miscellaneous Amy-2.1 and Amy-2.2 • IRS in the amylase promoter • Pancreas-specific • Amy-2.2 is stimulated by insulin and carbohydrate • Amy-2.1 is not responsive to either • 당뇨 쥐에서 amylase promoter의 30-bp fragment (from -167~-138) 의 activity 가 정 상 쥐보다 훨씬 떨어짐을 발견=> 이 부위가 IRS 31 Identification of Negative insulin response sequences • The Phosphoenolpyruvate Carboxykinase-like Motif – – – – Phosphoenolpyurvate Carboxykinase IGF binding protein-1 Tyrosine Aminotransferase Glucose-6-phosphatase catalytic subunit • Apolipoprotein CIII • Candidates for the insulin response factor – Models of insulin response Factor Action – Mechanisms of insulin signaling through the insulin response Factor • Miscellaneous 32 33 34 35