* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mol Bio CH 14 Nov 15
Citric acid cycle wikipedia , lookup
Western blot wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA silencing wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
Metalloprotein wikipedia , lookup
Messenger RNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene expression wikipedia , lookup
Peptide synthesis wikipedia , lookup
Proteolysis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
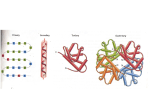
Goals: 1 - structure of messenger RNA and translation 2 - preparation of amino acids 3 - building a protein 4 components to protein synthesis (translation): 1 - mRNA Template 2 - transfer RNA (tRNA) bound to amino acids Substrate 3 - aminoacyl-tRNA sythetases (charge a.a.s) Substrate 4 - ribosomes Machinery mRNA can be translated 3 ways (3 frames) Many prokaryotic mRNAs have a Shine-Dalgarno sequence -Interacts with the ribsome to position next to start codon -Eukaryotic mRNAs may have a Kozak sequence - similar function -Other (less well understood) mechanisms function for mRNAs without these sequences Anatomy of tRNAs -All have same generic shape -Small differences allow specific tRNA to be matched with specific a.a.s “Charging” an amino acid requires ATP consumption -High-energy bond created between tRNA and a.a. -Energy used during protein synthesis -Different synthetase enzymes attach a.a. to tRNA at either the 2’ or 3’ OH of RNA Each amino acid has its own synthetase enzyme -RNA nucleotides can be modified -Clover structure results in nontraditional base pairing -Unique features of each tRNA are exploited (though not usually the anticodon) The ribosome is made of RNA and protein subunits Ribosomes are very large and complex Ribosomes assemble and disassemble with each translation event Peptide bond formation uses energy stored in the amino acid - tRNA high energy bond -Release of tRNA/formation of peptide bond is a 2 step process -The most recently added amino acid remains bound to tRNA 1 2 QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture.