* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Bottromycin wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Genetic code wikipedia , lookup
Molecular evolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Biochemistry wikipedia , lookup
List of types of proteins wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Proteolysis wikipedia , lookup
Structural alignment wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
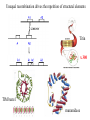
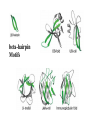
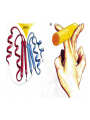
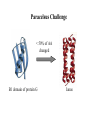
Modularity as an Organizing Principle in Protein Structure Unequal recombination drives the repetition of structural elements Titin x 300 Core of an average domain ~150 AA 20 different amino acids –> 20150 = 10195 different sequences Of these ~1038 are expected to have different fold (i.e. less than 20% sequence identity) Estimated number of naturally occurring folds ~1000 Fraction of theoretically possible “folds” used in nature ~ 1/1034 = 0.00000000000000000000000000000001% Super secondary Structure elements Unequal recombination drives the repetition of structural elements Titin x 300 TIM barrel muramidase Structures with alpha-hairpin motifs beta-hairpin Motifs beta-alpha-beta Motif QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Proteins with more than 30% AA identity almost always adopt the same fold. Stability -Gfolding Proteins as “Islands of Stability” in Sequence Space folded unfolded Sequence Bridges in between islands QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Cordes et al. Nat. Struct. Biol 2000 Dec;7(12):1129-32. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Glykos, N.M., Cesareni, G. & Kokkinidis, M. (1999), Structure 7, 597-603 Paracelsus Challenge < 50% of AA changed B1 domain of protein G Janus