* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Prediction of protein disorder: basic concepts and practical hints
Histone acetylation and deacetylation wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Gene expression wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Epitranscriptome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Silencer (genetics) wikipedia , lookup
List of types of proteins wikipedia , lookup
Immunoprecipitation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Drug design wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Proteolysis wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein adsorption wikipedia , lookup
Ligand binding assay wikipedia , lookup
Cooperative binding wikipedia , lookup
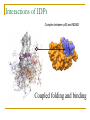
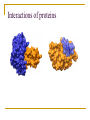
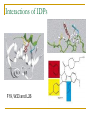
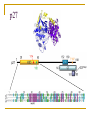
PART II. Prediction of functional regions within disordered proteins Zsuzsanna Dosztányi MTA-ELTE Momentum Bioinformatics Group Department of Biochemistry Eotvos Lorand University, Budapest, Hungary [email protected] LDR (40<) protein, % Protein disorder is prevalent 60 E 40 A 20 B 0 kingdom Protein disorder is functional Xie H et al. J Proteome Res. 2007, 6, 1882-1898 Protein disorder is important Prion protein Prion disease CFTR Cystic fibrosis t Alzheimer’s -synuclein Parkinson’s p53, BRCA1 cancer Functions of intrinsically disordered proteins I II III IV V Entropic chains Linkers Molecular recognition Protein modifications (e.g. phosphorylation) Assembly of large multiprotein complexes Interactions of IDPs Complex between p53 and MDM2 Coupled folding and binding Interactions of proteins Interactions of proteins Coupled folding and binding Functional advantages Weak transient, yet specific interactions Post-translational modifications Flexible binding regions that can overlap Evolutionary plasticity Signaling Regulation Coupled folding and binding Experimental difficulties Highly flexible Weak interactions Short half-life Complexes of IDPs in the PDB: ~ 200 Known instances: ~ 2,000 Estimated number of such interactions in the human proteome: ~ 100,000 Interactions of IDPs F19, W23 and L26 p27 Cyclin-dependent kinase (Cdk) inhibitor, p27Kip1 (p27) Cell cycle regulation Binds to cdk-cyclin komplex and inhibitis their activity Fully disordered protein p27 p27 Partial unfolding enables the phosphorylation of Tyr88, starting a series of signaling events that leads to the beginning of S phase. Prediction of functional regions within IDPs Disordered binding regions (ANCHOR) Linear motifs (ELM, SlimPred) Morfs (Morfpred) Specific conservation pattern Disordered protein complexes • Interaction sites are usually linear (consist of only 1 part) • enrichment of interaction prone amino acids Sequence No need for structure, binding sites can be predicted from sequence alone Complex between p53 and MDM2 Binding sites Prediction of disordered binding regions – ANCHOR What discriminates disordered binding regions? A cannot form enough favorable interactions with their sequential environment It is favorable for them to interact with a globular protein Based on simplified physical model Based on an energy estimation method using statistical potentials Captures sequential context ANCHOR Human p53 C –terminal region ANCHOR and linear motifs NCOA2 transcription co-activator The regions between 600-800 is disordered Contains 3 receptor binding motifs: xLxxLLx (LIG_NRBOX) LMs and Disordered Binding Regions Linear motifs and disordered binding regions often overlap Complementary approaches Prediction of disordered binding regions can help to increase likelihood of true instances LMs and Disordered Binding Regions Linear motifs and disordered binding regions often overlap Complementary approaches Prediction of disordered binding regions can help to increase likelihood of true instances Machine learning approaches SlimPred: trained on ELM database Morfpred: trained on short chains in complex Very small datasets Negative datasets Conservation Conservation patterns of linear motifs No evolutionary constraints to keep the structure Strong constraints on functional site Island-like conservation SlimPrints Generates sequence alignments of orthologous sequences Relative conservation score per position Filters out less reliable regions Fails if sequences are too divergent, or too similar http://bioware.ucd.ie/slimprints.html.