* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download lecture 5
Gene expression wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Signal transduction wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Peptide synthesis wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Expression vector wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Biochemistry wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Metalloprotein wikipedia , lookup
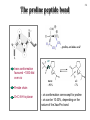
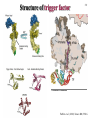
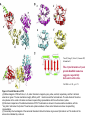
5-1 Protein folding catalysts Protein folding catalysts - peptidyl prolyl isomerases (PPIases) - protein disulfide isomerases (PDIases) 5-2 The proline peptide bond - proline, an imino acid 180º trans conformation favoured ~1000-fold over cis R=side chain O=C-N-H is planar trans ~93% cis ~7% - cis conformation rare except for proline - cis can be 10-30%, depending on the nature of the Xaa-Pro bond 5-3 Proline cis-trans isomerization - slow because it involves rotation about a partial double-bond (t1/2 between 10-100 sec at 25ºC) - cis-trans equilibria more common in flexible regions of native proteins (e.g., coils) OR: during protein folding partial double-bond character - strong acids favour cis-trans isomerization by protonating the nitrogen atom - proline residues disrupt alpha-helices; often found in turns - cis-trans isomerization could be used as a molecular switch + Catalysis of cis-trans isomerization - simple reaction; does not involve breaking or forming bonds - mechanism: catalysis by distortion and transition state containing partially-rotated C-N bond - this would result in a reduced partial double-bond character PPIase, Peptidyl Prolyl Isomerase, catalyzes proline cis-trans isomerization - active site of PPIase hydrophobic in character; conserved Arg residue of a PPI might be involved in H-bond formation with N:, producing C-N bond with more single-bond character + Peptidyl prolyl isomerases Three classes are known: • Cyclophilins - ubiquitous; 11 different ones found in S. cerevisiae; not essential for viability - binds cyclosporin A • FKBP binding proteins - no sequence similarity with cyclophilins; many different members found in eukaryotes, as well as prokaryotes; not essential for viability. Yeast mutant lacking all its cyclophilins and FKBP binding proteins still alive! - bind immunosuppressants FK506 and rapamycin (but not cyclosporin A) - both cyclophilins and FKBP’s form complexes with the molecular chaperone Hsp90, perhaps to catalyze cis-trans isomerization as well as to assist folding (or modulate protein conformation) • Parvulins - not related to cyclophilins or FKBP binding proteins, and are not inhibited by cyclosporin A, FK506 or rapamycin - occur as small proteins of <100 amino acids or as domains of larger proteins - have high PPIase activity .... and they differ in their activities and specificities, cellular localization, and binding partner(s) Implicated in: protein folding, protection against stress, apoptosis, cell cycle progression, etc. etc. 5-4 5-5 Assay methods for PPIases Chymotrypsin-coupled assay - chymotrypsin cleaves only the trans-form of the Xaa-Pro bond amino acid of a small model peptide such as N-succinyl-Ala-Xaa-Pro-Phe-p-nitroanilide - in aqueous solution, 90% of Xaa-Pro bond of this molecule is in trans-conformation - after addition of excess amount of chymotrypsin, the trans form of Xaa-Pro bond is cleaved instantaneously - hydrolysis rate of the remaining 10% Xaa-Pro bond is limited by its cis to trans isomerization - cis-trans isomerization rate of model peptide is measured by the release of p-nitroanilide spectrophotometrically Chymotrypsin-free assay - in a mixture of TFE and LiCl, the N-succinyl-AXPF-p-nitroanilide peptide is approximately 50% in the cis conformation - upon dilution in buffer, cis-trans isomerization occurs, decreasing cis content to ~10% - small differences in absorbance between the cis and trans forms of the prolyl imide bond in the model peptide are then measured at 330 nm NMR - could also monitor cis-trans isomerization by nuclear magnetic resonance (NMR), but this method is more expensive, slower, and requires more protein Chymotrypsin-coupled assay N-succinyl-Ala-Xaa-Pro-Phe-p-nitroanilide dilute in aqueous buffer N-succinyl-Ala-Xaa-ProPhe-p-nitroanilide N-succinyl-Ala-Xaa-ProPhe-p-nitroanilide 90% Xaa-Pro (trans) 10% Xaa-Pro (cis) add EXCESS chymotrypsin cis-trans isomerisation change in absorbance CLEAVED IMMEDIATELY time cleaved over time, depending on rate of isomerisation 5-6 Cyclophilins Discovery - 1989: an 18 kDa, cytosolic peptidyl prolyl isomerase from kidney was shown to be nearly identical to a cyclophilin known as Cyp A, a receptor protein for the immunosuppressant cyclosporin A Cyclosporin A - widely used to suppress graft rejection after organ transplantation - immunosuppressive because it inhibits synthesis of lymphokines (interleukins, macrophage colony-stimulating factor, etc.) - also has anti-viral, anti-fungal, anti-multi-drug resistance activities - binds very tightly to active site of Cyclophilin A; very hydrophobic cyclosporin A 5-7 Cyclophilin A with Gly-Pro GP peptide Arg Catalysis of cis-trans isomerization - simple reaction; does not involve breaking or forming bonds - mechanism: catalysis by distortion and transition state containing partially-rotated C-N bond - this would result in a reduced partial double-bond character - active site of Cyp A PPIase hydrophobic in character; conserved Arg residue of a PPI might be involved in H-bond formation with N:, producing C-N bond with more single-bond character - The pipecolic amide moiety of FK506, which probably mimics the proline residue of peptide or protein substrates, is bound in a hydrophobic pocket of FKBP, presumably at the active site FK506 rapamycin GP peptide Arg cyclophilin A in complex with dipeptide, GP - R55A mutant has <0.1% activity 5-8 FKBP binding protein: trigger factor - trigger factor (TF), a 48 kDa protein with FKBP domain, occurs only in bacteria - most efficient PPIase found to date - tightly associated with the ribosome; some of it is cytosolic - not essential for E. coli viability but synthetic lethality with Hsp70 (DnaK) chaperone - can be cross-linked to nascent polypeptides Domain structure: N C ribosomebinding domain FKBP domain peptidebinding domain - can be cross-linked to nascent polypeptides - prepare E. coli in vitro translation extract, translate mRNA in the presence of N-5-azido-2nitrobenzoyl)-Lys-tRNALys; photocross-link aryl azide group; immunoprecipitate protein N3 Lys-N(epsilon)-NHCONO2 - cross-links non-specifically to proteins - the photo-activated probe is made in the lab - works in rabbit reticulocyte lysate translations as well Structure of trigger factor 5-9 TF bound to ribosome Ferbitz et al. (2004) Nature 431, 590-6. Protein disulfide isomerases - PDIases belong to a large, ubiquitous group of proteins possessing the thioredoxin fold - there is little sequence similarity between the different proteins - overall enzymatic activity: formation of correct disulfide bonds - formation (I) and reduction (II) of disulfide bonds (III), ‘unscrambling’ of incorrect disulfide bonds forming breaking correcting Notes: - not all members of this large family can catalyze disulfide bond isomerization - substrate specificity differs between different members - LOCALIZATION: - the cytoplasm has a reducing environment where disulfide bonds do not occur • disulfide-bonded proteins occur in the endoplasmic reticulum in eukaryotes; secreted proteins • periplasmic space in prokaryotes 5-10 5-11 Thioredoxin fold - catalytic region of the thioredoxin domain contains an Cys-X-X-Cys motif; most contain 1, but PDI from the ER has 2 of these motifs • formation of disulfide bond requires oxidation of the cysteine sulfhydryls • The redox potential of a molecule is the ability of an electron donor to reduce an electron acceptor. A higher reduction (redox) potential means that a molecule will be a better oxidant • The sequence of the two amino acids found between the cysteines in a CXXC active site influences the redox potential of the enzyme. Thioredoxin (CPYC), PDI (CGHC), and DsbA (CPHC) have redox potentials of -270 mV, -147 to -175 mV, and -122 mV (i.e., thioredoxin is the best reductant, DsbA is the best oxidant) Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 2006 vol. 124, p. 61-73. Figure 2. Overall Structure of PDI (A) Ribbon diagram of PDI with the a, b, b′, and a′ domains in magenta, cyan, yellow, and red, respectively, and the C-terminal extension in green. The two orientations roughly differ by a 90° rotation around the horizontal axis. The side chains of the active site cysteines in the a and a′ domains are shown in space-filling representation with the sulfur atoms in yellow. (B) Structural comparison of the individual domains of PDI. The domains are shown in the same relative orientation, with the “long helix” side below the β sheet. The active-site cysteine residues in the a and a′ domains are shown in space-filling representation. (C) Secondary structure diagram of the canonical thioredoxin fold with α helices in green and β strands in red. The location of the active site is indicated by a red oval. 5-12 Other oxidizing/reducing agents in vivo - Glutathione is an abundant redox buffer in the endoplasmic reticulum, but it is not essential in vivo - it undergoes the following redox (oxidation/reduction) cycle: GSH + S-S GSSG + SH HS - 2:1 ratio of GSH:GSSG in ER (optimum ratio for rearrangements) - 100:1 ratio in cytosol in vitro - dithiothreitol (DTT) is a useful chemical for disulfide bond reduction OH HS HO SH DTT OH HS 2-mercaptoethanol