* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Restriction enzyme wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Photosynthesis wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Paracrine signalling wikipedia , lookup
Metalloprotein wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Electron transport chain wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Catalytic triad wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Biochemical cascade wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Citric acid cycle wikipedia , lookup
Biosynthesis wikipedia , lookup
Microbial metabolism wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Biochemistry wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
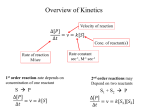
Effect of environment on enzyme activity Substrate concentration pH Temperature Substrate concentration Enzyme activity increases with increasing substrate concentrations At a certain concentration the enzyme will be saturated and operate at maximum velocity = Vmax Substrate concentration Plot of velocity (activity) vs. substrate concentration results in a hyperbola Michaelis constant (Km) Used to measure the affinity of an enzyme for its substrate Km = substrate concentration required to achieve half maximal velocity Effect of pH and temperature on enzyme activity Enzymes are most active at optimum pH and temperatures Deviations from the optima can slow activity and damage the enzyme Effect of pH and temperature on enzyme activity Loss of activity due to extreme pH, temperature or other factors is known to as denaturation Temperature and pH optima of microorganism’s enzymes usually reflect the microorganism’s environment Enzyme inhibition Many poisons and antimicrobial agents are enzyme inhibitors Can be accomplished by competitive or noncompetitive inhibitors Competitive inhibitors - compete with substrate for the active site Noncompetitive inhibitors - bind at another location Competitive inhibitors Usually resemble the substrate but cannot be converted to products Noncompetitive inhibitors Bind to the enzyme at some location other than the active site Do not compete with substrate for the active site Binding alters enzyme shape and slows or inactivates the enzyme Heavy metals often act as noncompetitive inhibitors (e.g. Mercury) Metabolic regulation Important to conserve energy and resources Cell must be able to respond to changes in the environment Changes in available nutrients will result in changes in metabolic pathways Metabolic regulation Metabolic channeling Stimulation or inhibition of enzyme activity Transcriptional regulation of enzyme production Allosteric enzymes Activity of enzymes are altered by small molecules known as effectors or modulators Effectors bind reversibly and noncovalently to the regulatory site Binding alters the conformation of the enzyme Allosteric enzymes Positive effectors increase activity Negative effectors decrease activity ACTase regulation Regulation of aspartate carbamyltransferase is a well studied example of allosteric regulation CTP inhibits activity and ATP stimulates activity ACTase regulation Binding of effectors cause conformational changes that result in more or less active forms of the enzyme ACTase regulation Binding of substrate also increases enzyme activity (more than one active site) Velocity vs. substrate curve is sigmoid Covalent modification of enzymes Attachment of group to enzyme can result in stimulation or inhibition of activity Attachment is covalent and reversible Covalent modification of enzymes Attachment of phosphate groups often used to regulate enzyme activity Other groups can also be used to regulate enzyme activity Feedback inhibition Metabolic pathways contain at least one pacemaker enzyme Usually catalyzes the first reaction in the pathway Activity of the enzyme determines the activity of the entire pathway Feedback inhibition Feedback inhibition occurs when the end product interacts with the pacemaker enzyme to inhibit its activity Branching pathways regulate enzymes at branch points Overview of metabolism Metabolism = the total of all chemical reactions occurring within the cell Catabolism = the breaking down of complex molecules into simple molecules with the release of energy Anabolism = the synthesis of complex molecules from simple molecules with the use of energy Sources of energy Microorganisms use one of three sources of energy Phototrophs - radiant energy of the sun Chemoorganotrophs oxidation of organic molecules Chemolithotrophs oxidation of inorganic molecules Electron acceptors Chemotrophs vary regarding their final electron acceptors Fermentation - no exogenous electron acceptor is required Aerobic respiration - oxygen is the final electron acceptor Anaerobic respiration - another inorganic molecule is acceptor Electron acceptors Chemolithotrophs can use oxygen or another inorganic molecule as the final electron acceptor The three stages of catabolism Catabolism can be broken down into three stages Stage 1 Larger molecules (proteins, polysaccharides, lipids) are broken down into their constituents Little or no energy is generated The three stages of catabolism Stage 2 Amino acids, monosaccharides, fatty acids, glycerol and other products degraded to a few simpler products Can operate aerobically or anaerobically Generates some ATP and NADH or FADH The three stages of catabolism Stage 3 Nutrient carbon is fed into the tricarboxylic pathway and oxidized to CO2 ATP, NADH and FADH produced ATP generated from oxidation of NADH and FADH in electron transport chain Amphibolic pathways Pathways that can function both catabolically and anabolically Glycolysis and the tricarboxylic acid cycle are two of the most important amphibolic pathways Most reactions are reversible The glycolytic pathway/glycolysis Also known as the EmbdenMeyerhof pathway Most common pathway of degradation of glucose to pyruvate Found in all major groups of microorganisms Can function aerobically or anaerobically The glycolytic pathway/glycolysis Occurs in 2 stages The six-carbon stage Glucose is phosphorylated 2x and converted to fructose-1,6-bisphosphate Other sugars converted to glucose-6-phosphate or fructose-6-phosphate and fed into pathway The glycolytic pathway/glycolysis The six-carbon stage Does not yield energy Uses 2 ATPs Serves to “prime the pump” The glycolytic pathway/glycolysis The three-carbon stage Fructose-1,6-bisphosphate split in half by fructose-1,6bisphosphate aldolase Yields glyceraldehyde-3phosphate and dihydroxyacetone phosphate The glycolytic pathway/glycolysis The three-carbon stage Dihydroxyacetone phosphate readily converts to glyceraldhyde3-phosphate Fructose-1,6-bisphosphate 2 glyceraldehyde-3-phosphate The glycolytic pathway/glycolysis The three-carbon stage Glyceraldehyde-3-phosphate converted into pyruvate in 5 steps Oxidized by NAD+ and a phosphate is added 1,3-bisphosphoglycerate Phosphate on carbon 1 donated to ADP to form ATP Substrate-level phosphorylation The glycolytic pathway/glycolysis The three-carbon stage 3-phosphoglycerate shifted to carbon 2 2-phosphoglycerate Dehydration results in high energy phosphate bond in phosphoenolpyruvate Phosphate transferred to ADP to form ATP (substrate-level phosphorylation) The glycolytic pathway/glycolysis Glucose 2 pyruvates + ATP +NADH 2 ATP used in six-carbon stage 4 ATP + 2 NADH formed in threecarbon stage The glycolytic pathway/glycolysis Glucose + 2ADP + Pi + 2NAD+ 2 pyruvate + 2ATP + 2NADH