* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Protein Metabolism - Orange Coast College
Peptide synthesis wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Western blot wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Gene expression wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Messenger RNA wikipedia , lookup
De novo protein synthesis theory of memory formation wikipedia , lookup
Proteolysis wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
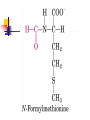
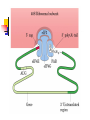
Protein Metabolism Protein Synthesis Protein Synthesis: an overview PS proceeds from N -terminus to Cterminus (amino to carboxyl) Ribosomes:read mRNA in 5’ to 3’ direction Polyribosomes (polysomes) Max. density ~ 1 ribosome/80 nucleotides Protein Synthesis: an overview Chain elongation: links the growing chain to incoming tRNA’s AA residue AA residues: added to C-terminus Ribosome (prokaryote) has 3 tRNA binding sites The P site: peptidyl site Binds peptidyl –tRNA Peptidyl-tRNA holds polypeptide After peptide bond formation, binds deacylated tRNA The A site: amino-acyl site Binds incoming aminoacyl-tRNA Protein Synthesis: an overview Ribosome (prokaryote) has 3 tRNA binding sites After peptide bond formation Deacylated tRNA released from P site Replaced in P site by newly formed peptidyl-tRNA A site is vacated The E site: exit site Recent finding Largely confined to 50S subunit Deacylated tRNA dissociate from the ribosome Protein Synthesis: an overview Five (5) major stages of protein synthesis 1.Activation of Amino Acids 20 AA 20 aminoacyl-tRNA synthetases 20 or more tRNAs (min 32) ATP Mg2+ Protein Synthesis: an overview Five (5) major stages of protein synthesis 2.Initiation mRNA N-Formylmethionyl-tRNA Initiation codon in mRNA (AUG) Ribosome: 30S and 50S subunit Initiation factors: IF-1. IF-2, IF-3 GTP Mg2+ Protein Synthesis: an overview Five (5) major stages of protein synthesis 3.Elongation Initiation complex (functional 70S ribosome) aminoacyl-tRNAs specified by codons Peptidyl transferases Elongation factors: EF-Tu. EF-Ts, EF-G GTP Mg2+ Protein Synthesis: an overview Five (5) major stages of protein synthesis 4. Termination and Release Termination codon in mRNA Polypeptide releasing factor: RF1, RF2, RF3 ATP 5. Folding and Processing Enzymes (lots!) Cofactors (lots!) P S: stage one; activation of AA Takes place in the cytosol Each AA is attached to specific tRNA ATP to AMP + Ppi Catalyzed by MG 2+ -depending aminoacyltRNA synthetases Aminoacylated tRNA is said to be charged P S: stage two; Initiation Initiation codon recognized by tRNAfmet N-formylmethionine residue (fmet) Special tRNA Special aminoacyl-tRNA synthetase First: bind tRNAfmet with Met Then: N –formulates the Met residue Note: proteins are post-translationally modified Deformylation of fmet residue Sometimes: removal of N-terminal Met P S: stage two; Initiation Initiation codon recognized by tRNAfmet In eukaryotes: All polypeptides synthesized by cytoplasmic ribosomes Begin with Met residues (not Have special initiating tRNA met) f Mitochondrial/chloroplast products are like polypeptides P S: stage two; Initiation The Shine-Dalgarno Sequence: John Shine and Lynn Dalgarno in 1974 An initiating signal in mRNA Base pair with (antiparallel): 8-13 bp to the 5’ side of initiation codon 4-9 purine residues Complementary pyrimidine-rich sequence Nearnear 3’ end of 16SrRNA on 30S subunit mRNA-rRNA interactions Sets mRNA in the correct position Initiation of transcription P S: stage two; Initiation Three Stages: assemble Initiation Complex Requires Initiation factors Not permanently associated with ribosome 3 (in E. coli ): IF-1, IF-2, IF-3 Stage one (1): 30S subunit binds IF-3, IF-1 Prevents premature joining of LG and SM subunits mRNA binds to 30S Initiation codon (AUG) to P-site on 30S subunit Guided by Shine-Delgarno sequence P S: stage two; Initiation Three Stages: assemble Initiation Complex Stage two (2): 30S subunit with IF-3 with mRNA binds w/IF-2 IF-2 is already bound to: GTP fMet-tRNA fmet Anticodon and codon pair P S: stage two; Initiation Three Stages: assemble Initiation Complex Stage three (3): Large complex formed in stage 2 combines with 50S subunit GTP to GDP and Pi IF-3, IF-1, IF-2 are released IF-3 released before 50S attaches irreversible P S: stage two; Initiation Three Stages: assemble Initiation Complex At end of Initiation: fMet-tRNA fmet with mRNA with Ribosome complex is formed fMet-tRNA fmet in P site A site ready Eukaryotic Initiation is similar More initiation factors No Shine-Dalgarno Cap is located; the 1st AUG downstream P S: stage three; Elongation 3 stage cycle that is repeated Adds AA to C-terminus Up to 40 residues/sec Elongation factors (EF) P S: stage three; Elongation Aminoacyl-tRNA binding GTP with EF-Tu with aminoacyl-tRNA binds to ribosome aminoacyl-tRNA: bound in codon-anticodon interaction at A site GTP to GDP + Pi EF-Tu to GDP + Pi are released Regenerate GTP EF-Tu with GDP + EF-T3 to EF-Tu with EF-Ts + GDP EF-Tu with EF-Ts + GTP to EF-Tu with GTP + EF-Ts P S: stage three; Elongation Transpeptidation: Peptide bond formation Transfer of N-formylmethionyl group From tRNA in P site To amino group of 2nd AA in A site Forms a dipeptidyl-tRNA in A site tRNA fmet in P site Peptidyl transferases Catalyzes bond formation on LG subunit Catalyzed by 23S rRNA (Harry Noller, 1992) P S: stage three; Elongation Translocation: Ribosomes moves toward 3’ end by one codon Shift requires Dipeptidyl-tRNA moves to Psite Deacylated- tRNA fmet released New codon (3rd) into A site EF-G (translocase) GTP Believed to be accompanied by 3-D changes in ribosome P S: stage three; Elongation Repeat elongation cycle Need 2 GTP for each added AA residue Protein chain always remains attached to a tRNA P S: stage four; TERMINATION Signaled by termination codon When termination codon is in the A site 3 releasing factors RF1, RF2, RF3 Hydrolysis of terminal peptidyl-tRNA bond Release of protein and last tRNA Dissociation of ribosome P S: stage four; TERMINATION When termination codon is in the A site 3 releasing factors RF1: reacts to UAG, UAA RF2: reacts to UGA, UAA RF3: ? RF bind at termination codon Peptidyl transferase gives chain to H2O Eukaryotes One RF: eRF Recognizes all 3 termination codons P S: stage five; post-translational modification To become mature, polypeptides must fold to native conformations Disulfide bonds must form Multisubunit proteins: subunits must combine Must be modified by enzymes Proteolytic cleavage Most common P-T modification Eg: all proteins have fMet residue removed Eg: Conversion of trypsinogen to trypsin P S: stage five; post-translational modification Covalent modifications E.g.: Methylations Hydroxylations Etc Changes in functional groups and radical groups