* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download GLYPHOSATE RESISTANCE Background / Problem
Artificial gene synthesis wikipedia , lookup
History of genetic engineering wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Minimal genome wikipedia , lookup
Genome (book) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Human genetic variation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Gene expression programming wikipedia , lookup
Point mutation wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
The Selfish Gene wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genetic drift wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Group selection wikipedia , lookup
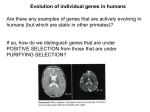
Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium November 12, 2012 Last Time Sequence data and quantification of variation Infinite sites model Nucleotide diversity (π) Sequence-based tests of neutrality Tajima’s D Hudson-Kreitman-Aguade Synonymous versus Nonsynonymous substitutions McDonald-Kreitman Today Signatures of selection based on synonymous and nonsynonymous substitutions Multiple loci and independent segregation Estimating linkage disequilibrium Using Synonymous Substitutions to Control for Factors Other Than Selection dN/dS or Ka/Ks Ratios Types of Mutations (Polymorphisms) Synonymous versus Nonsynonymous SNP First and second position SNP often changes amino acid UCA, UCU, UCG, and UCC all code for Serine Third position SNP often synonymous Majority of positions are nonsynonymous Not all amino acid changes affect fitness: allozymes Synonymous & Nonsynonymous Substitutions Synonymous substitution rate can be used to set neutral expectation for nonsynonymous rate dS is the relative rate of synonymous mutations per synonymous site dN is the relative rate of nonsynonymous mutations per non-synonymous site = dN/dS If = 1, neutral selection If < 1, purifying selection If > 1, positive Darwinian selection For human genes, ≈ 0.1 Complications in Estimating dN/dS Multiple mutations in a codon CGT(Arg)->AGA(Arg) give multiple possible paths Two types of nucleotide base substitutions resulting in SNPs: transitions and transversions not equally likely CGT(Arg)->AGT(Ser)->AGA(Arg) CGT(Arg)->CGA(Arg)->AGA(Arg) Back-mutations are invisible Complex evolutionary models using likelihood and Bayesian approaches must be used to estimate dN/dS (also called KA/KS or KN/KS depending on method) (PAML package) http://www.mun.ca/biology/scarr/Transitions_vs_Transversions.html dn/ds ratios for 363 mouse-rat comparisons Most genes show purifying selection (dN/dS < 1) Some evidence of positive selection, especially in genes related to immune system interleukin-3: mast cells and bone marrow cells in immune system Hartl and Clark 2007 McDonald-Kreitman Test Conceptually similar to HKA test Uses only one gene Contrasts ratios of synonymous divergence and polymorphism to rates of nonsynonymous divergence and polymorphism Gene provides internal control for evolution rates and demography Application of McDonaldKreitman Test: Aligned 11,624 gene sequences between human and chimp Calculated synonymous and nonsynonymous substitutions between species (Divergence) and within humans (SNPs) Identified 304 genes showing evidence of positive selection (blue) and 814 genes showing purifying selection (red) in humans Positive selection: defense/immunity, apoptosis, sensory perception, and transcription factors Purifying selection: structural and housekeeping genes Bustamente et al. 2005. Nature 437, 1153-1157 Genes showing purifying (red) or positive (blue) selection in the human genome based on the McDonald-Kreitman Test Bustamente et al. 2005. Nature 437, 1153-1157 How can you differentiate between effects of selection and demographic effects on sequence variation? Will this work for organellar DNA? Extending to Multiple Loci So far, only considering dynamics of alleles at single loci Loci occur on chromosomes, linked to other loci! “The fitness of a single locus ripped from its interactive context is about as relevant to real problems of evolutionary genetics as the study of the psychology of individuals isolated from their social context is to an understanding of man’s sociopolitical evolution” Richard Lewontin (quoted in Hedrick 2005) Size of region that must be considered depends on Linkage Disequilibrium Gametic (Linkage) Disequilibrium (LD) Nonrandom association of alleles at different loci into gametes Haplotype: Genotype of a group of closely linked loci LD is a major factor in evolution LD itself provides insights into population history Estimation of LD is critical for ALL population genetic data Nomenclature and concepts Two loci, two alleles Frequency of allele i at locus 1 is pi Frequency of allele i at locus 2 is qi p1 A1 B1 q1 p2 A2 B2 q2 n n p q i 1 i i 1 i 1 Nomenclature and concepts Genotype is written as A1B1 A2B2 A1 B1 A2 B2 A1 and B1 are in coupling phase A1 and B2 are in repulsion phase Gametic Disequilibrium Easiest to think about physically linked loci, but not necessarily the case A1B1 A2B2 Meiosis A1B1 A1B2 A2B1 A2B2 What Are of Gametes in a p1q1 Expected p1q2 Frequencies p2q1 p2q2 Population Under Independent Assortment? What are expected frequency of Gametes with complete linkage? p1 A1 B1 q1 p2 A2 B2 q2 A1B1 A2B2 Meiosis A1B1 x11 A1B2 A2B1 A2B2 x12 x21 x22 Linkage disequilibrium measure, D Independent Assortment: With LD: Substituting from above table: D x11x22 x12 x21 Problem: D is sensitive to allele frequencies Can’t have negative gamete frequencies Maximum D set by allele frequencies Solution: D' = D/Dmax ranges from -1 to 1 Example, if D is positive: p1=0.5, q2=0.5, Dmax=0.25 but p1=0.1, q2=0.9, Dmax=0.09 Dmax Calculation: If D is positive, Dmax is lesser of p1q2 or p2q1 If D is negative, Dmax is lesser of p1q1 or p2q2 LD can also be estimated as correlation between alleles r 2 D p1 p2 q1q2 r can also be standardized to a -1 to 1 scale It is equivalent to D’ in this case r' D p1 p2 q1q2 D' Dmax p1 p2 q1q2 Recombination Shuffling of parental alleles during meiosis A1B1 A2B2 A1 B1 A1 B2 A2 B2 A2 B1 Occurs for unlinked loci and linked loci Rate of recombination for linked markers is partially a function of physical distance What is the expected recombination rate for unlinked loci? A1B1 A2B2 Meiosis A1B1 Coupling nr c nr nc A1B2 A2B1 A2B2 Repulsion Repulsion Coupling Where nr is number of repulsion phase gametes, and nc is number of coupling phase gametes LD is partially a function of recombination rate Expected proportions of gametes produced by various genotypes over two generations First generation Where c is the recombination rate and D0 is the initial amount of LD (Second generation) Recombination degrades LD over time D1 x'11 x'22 x'12 x'21 ( x11 cD0 )( x22 cD0 ) ( x12 cD0 )( x21 cD0 ) D1 (1 c) D0 Dt (1 c) D0 t ct Dt e D0 Where t is time (in generations) and e is base of natural log (2.718) Effects of recombination rate on LD Decline in LD over time with different theoretical recombination rates (c) Even with independent segregation (c=0.5), multiple generations required to break up allelic associations Genome-wide linkage disequilibrium can be caused by demographic factors (more later)