* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Cell Bio/Physio Lecture 6 Objectives Sunday, August 14, 2011 11:41
Artificial gene synthesis wikipedia , lookup
Paracrine signalling wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Expression vector wikipedia , lookup
Gene expression wikipedia , lookup
Signal transduction wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Interactome wikipedia , lookup
Protein purification wikipedia , lookup
Biosynthesis wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
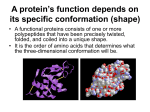
Cell Bio/Physio Lecture 6 Objectives Sunday, August 14, 2011 11:41 PM List the different types of proteins, including fibrous, globular, integral and peripheral, and describe their basic characteristics. Protein Description Example Location Fibrous Usually not soluble; formed by spiralshaped polypeptide molecules becoming linked together in cross-bridges. They have a rope like structure 1. 2. 3. 4. 1. Walls of large arteries 2. Hair 3. Muscles 4. Bones and tendons/ ligaments Globular Also known as spheroproteins comprising "globe"-like proteins that are more or less soluble in aqueous solutions (where they form colloidal solutions) 1. Enzymes- Biological catalysts which have an exposed active site They speed up biochemical reactions within the body 2. Hormones- Chemical messengers in the body. They have an effect on the animal's growth and metabolizm 3. Antibodies- Y-shaped and made by WBC called lymphocytes and their main purpose is to defend the body against antigens Integral Span the lipid bilayer; facilitate the transport of specific compounds by energy-requiring active transport, facilitate diffusion, or by forming pores of gated channels. Peripheral Bind the surface of the bilayer Elastin (Strong/elastic) Keratine- Strong/inelastic Actin & Myosin (Contractile) Collagen- Strong/inelastic Plasma membrane Define protein primary structure, and explain how joining amino acids into linear chains form proteins. Primary structure of a protein is the linear sequence of amino acids in the polypeptide chain. A condensation reaction joins amino acids together by amide linkage, water is released When peptide bonds are broken, it is done via hydrolysis reaction Describe the chemical properties of the peptide bond in terms of which bonds have partial double bond character and which are true single bonds. Relate these properties to bond rotations. Peptide bonds: Assumes a trans configuration in which successive alpha-carbons and their R groups are located on opposite sides of the peptide bond. Hybrid of two resonance structures- Double-bond character, so that the carboxyl and amide groups that form the bond must remain planar; Single bonds allow for rotation of atoms/groups between the alpha-carbon and the alpha-amino group and around the bong between the alphacarbon and the carbonyl group (Subject to steric contraints that maximize the distance between atoms in the different amino acid side chains and prohibit torsion angles that place the sidechain atoms too close to each other. Because of the resonance nature of the peptide bond, the C and N of the peptide bonds form a series of rigid planes. Rotation within allowed torsion angles can occur around the bonds attached to the alpha-carbon. The side chains are trans to each other, and alternate above and below the peptide chain. The actual peptide is a hybrid between the resonance forms shown, resulting in a partial negative charge on the carbonyl oxygen, a partial positive charge on the nitrogen, and partial double-bond character for the peptide bond itself. Explain the nomenclature for proteins and provide the name of a short peptide using three letter and single letter abbreviations. By Convention, the N-terminus is on the left. Naming begins with the N-terminus Peptides are named using the 3 letter abbreviations, whereas protein sequences use the 1 letter code Proteins go by common names, rather than by chemical names. Nonpolar, aliphatic: Glycine (gly), Alanine (ala), Proline (pro), Valine (val), Leucine (leu), Isoleucine (ile) Aromatic: Phenylalanine (phe), Tyrosine (tyr), Tryptophan (trp) Polar, uncharged: Asparagine (asn), Glutamine (gln), Serine (ser), Threonine (thr) Sulfur-containing: Methionine (met), Cysteine (cys) Charged: Aspartate (asp), Glutamate (glu), Arginine (arg), Lysine (lys), Histidine (his) List and define the factors that control protein folding. The primary structure of a protein dictates the way that it folds into its tertiary structure, which is a stable conformation that is identical to the shape of other molecules of the same protein (i.e. its native conformation) Chaperonins act as templates to overcome the kinetic barrier to reach a stable conformation. Prion proteins cause neurodegenerative diseases by acting as a template for misfolding Heat, acid, and other agents cause proteins to denature, that is, to unfold or refold and lose their native 3D conformation Define protein secondary structure. Regions within polypeptide chains form recurring, localized structures known as secondary structure: Alpha-Helix: Globular proteins, membrane-spanning domains, and DNA-binding proteins; Rigid stable conformation that maximizes hydrogen bonding while maintaining the allowed rotation angles of the polypeptide bonds. o Linear chain of amino acids wraps itself into a coil so that the amine hydrogen forms a hydrogen bond with the adjacent carboxyl oxygen: these H-bonds are the primary weak force that stabilized the alpha-helical structure o Amino acid side chains: Each oxygen of a carbonyl group of a peptide bond forms a hydrogen bond with the hydrogen atom attached to a nitrogen atom in a peptide bond 4 amino acids farther along the chain. The side chains jut out from the helix; Steric hindrance occurs if they come within their van der Waals radii of each other, and a stable helix cannot form. Beta-Sheets: Maximizes hydrogen bonding between the peptide backbones while maintaining the allowed torsion angles. The amino acid side chains of each polypeptide stand alternate between extending above and below the plane of the beta-sheet. Described as parallel if the polypeptide strands run in the same direction and antiparallel if they run in opposite directions. o Linear chain of amino acid aligns itself side-to-side that the amine hydrogen forms a hydrogen bond with the adjacent carboxyl oxygen: these H-bonds are the primary weak force that stabilizes the beta-sheet structure Describe the properties of an α-helix, including the orientation of the amino acid side chains. See above Describe the properties of a β-sheet, including the orientation of the amino acid side chains. Define the difference between parallel and anti-parallel β-sheets. See above Describe how the weak forces drive protein folding Hydrogen bonds Hydrophobic interactions Van der Waals Ionic interaction Define super-secondary protein structures, motifs and domains. Explain the role of domains in terms of the functional components of proteins Motifs: Each individual protein has a segment that binds to the DNA molecules, and a different segment that binds to its partner protein; DNA binding is highly specific; The geometry of the amino acid sequence matches exactly the geometry of the DNA sequence, allowing for this specific binding. Zinc finger- Consists of an alpha-helix and a beta-sheet in which 4 cystein and/or histidine residues coordinately bind a zinc ion. The nucleotide recognition signal of at least one zinc finger binds to a specific sequence of bases in the major groove of DNA Leucine zipper- Form from 2 distinct polypeptide chains. Each polypeptide contains a helical region in which leucine resides are exposed on one side. These leucines form hydrophobic interactions with each other, causing dimerization. The remaining helices interact with DNA Helix-turn-helix- Contain 3 helical regions, one of which brings to DNA, whereas the other lie on top and stabilize the interaction Helix-loop-helix- Contain helical regions that bind to DNA like leucine zippers. However, their dimerization domains consist of two helices, each of which is connected to its NDA-binding helix by a loop Domains: Usually form a structural or functional component of the proteins: Catalytic site Interaction site for binding other proteins Ligand binding site o Examples: Ca2+ transporting ATPase enzyme of muscle; ATP phosphorylates the enzyme, driving conformational changes; These conformational changes push Ca2+ across the membrane; Define protein tertiary structure Folding pattern of the secondary structural elements into a 3D conformation. Protein is fully functional in this form. The 3D structure is flexible and dynamic, with rapidly fluctuating movement in the exact position of amino acid side chains and domains. Maintains residues on the surface appropriately for the proteins cellular location, polar residues for systolic proteins, and hydrophobic residues for transmembrane proteins. Explain the process of protein denaturation and renaturation Define protein quaternary structure and explain the functional properties of this structure Apart from covalent disulfide bonds, proteins are held together by the weak forced These weak interactions can be overcome by environmental conditions such as: salt or charged compounds, pH, temperature, and organic solvents Cause protein to unfold Disulfide bonds broken only by reduction of S-S bond (They do not spontaneously reform) For many proteins, simply return the protein to a conducive environment, and it will refold to its native state- provided disulfide bonds are not broken Assembly of globular polypeptide subunits into a multisubunit complex can provide the opportunity for cooperative binding of ligands (i.e. O2 binding to hemoglobin), form binding sites for complex molecules (i.e. antigen binding to immunoglobin), and increase stability of the protein. The polypeptide chains of fibrous proteins such as collagen are aligned along an axis, have repeating elements, and are extensively linked to each other through hydrogen bonds. Refers to the association of individual polypeptide chain subunits in a geometrically and stoichiometrically specific manner. The prefixes homo and hetero are used to describe identical or different subunits. Protomer is a unit structure composed of nonidentical subunits List and describe the common protein modifications found in cells. Some proteins are modified after they are synthesized; Specific amino acids are modified at specific location on the protein. Important for localizing proteins to specific places in cell, thereby ensuring protein function at the place Can modify protein function for regulation purposes (Turn a protein on or off) o Examples: Glycosylation- Addition of small or large carbohydrate chains. Targets protein to outer surface of plasma membrane or for excretion. Attached to an oxygen atom or a nitrogen atoms of specific amino acids Fatty acylation or prenylation (lipid anchors)- Addition of a fatty acid, prenyl group, or a whole lipid molecule. Targets protein to membrane surfaces. Localized protein function to these places Regulatory modifications- Phosphorylation, acetylation or ADP-ribosylation or specific amino acids. Induces a conformation change that changed the activity of the protein. Reversible- Can be added and removed to reversibly regulate protein function Oxidized amino acids- Facilitate cross-linking or attachment.