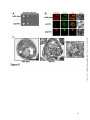
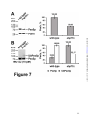
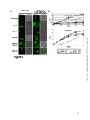
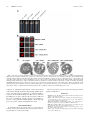
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Charakterisierung peroxisomaler und Lipid
Organ-on-a-chip wikipedia , lookup
Extracellular matrix wikipedia , lookup
Lipid bilayer wikipedia , lookup
Cytokinesis wikipedia , lookup
Theories of general anaesthetic action wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Model lipid bilayer wikipedia , lookup
Magnesium transporter wikipedia , lookup
Cell membrane wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Signal transduction wikipedia , lookup
Endomembrane system wikipedia , lookup
Charakterisierung peroxisomaler und Lipid-Droplet assoziierter Proteine der Hefe Saccharomyces cerevisiae Dissertation zur Erlangung des Grades eines Doktors der Philosophie der Fakultät für Biologie und Biotechnologie der Ruhr-Universität Bochum Internationalen Graduiertenschule Biowissenschaften der Ruhr-Universität Bochum Abteilung für Systembiochemie vorgelegt von Dipl.-Biol. & Biochem. Mykhaylo O. Debelyy aus Dnepropetrovsk, Ukraine Bochum Juli, 2011 Zusammenfassung In der vorliegenden Arbeit wurden Peroxisomen und Lipid-Droplet assoziierte Proteine der Hefe S. cerevisiae untersucht. Lpx1p und Ldh1p sind putative Hydrolasen und/oder Lipasen von Peroxisomen beziehungsweise Lipid-Droplets; Pex1p und Pex6p sind peroxisomale AAA Proteine und Ubp15p stellt ein deubiquitinilierendes Enzym dar. Es konnte gezeigt werden, dass Lpx1p in Peroxisomen lokalisiert ist, wo hingegen Ldh1p überwiegend zu Lipid-Droplets dirigiert wird. Lpx1p wie auch Ldh1p besitzen das für Lipasen der a/ß-Hydrolase-Familie typische Sequenzmotiv GXSXG. Beide Proteine tragen ein putatives, peroxisomales Typ 1 targeting Signal (PTS1) und weisen untereinander zwei homologe Bereiche auf. Während gezeigt werden konnte, das es sich bei Lpx1p um ein peroxisomales Enzym handelt, wurde anhand subzellulärer Lokalisationsstudien für Ldh1p eine überwiegende Lokalisation an Lipid-Droplets dargelegt. Für Lpx1p konnte ferner gezeigt werden, das dessen Lokalisation vom PTS1Rezeptor Pex5p abhängt. Ferner konnte für Lpx1p ein Huckepack-Transport in die Peroxisomen gezeigt werden. Dem gegenüber ist der Transport von Ldh1p zu den LipidDroplets unabhängig von dessen PTS1. Für Lpx1p und Ldh1p konnte in vitro mittels rekombinanter Proteine eine Triacylglycerol-Lipase wie auch–hydrolase Aktivität belegt werden. Es konnte gezeigt werden, dass Lpx1p nicht für das Vorhandensein funktioneller Peroxisomen benötigt wird, ein Umstand der eher auf eine metabolische als auf eine Biogenesefunktion des Proteins hinweist. Ldh1p ist hingegen notwendig für die Aufrechterhaltung normaler Konzentrationen an nicht-polaren und polaren Lipiden in den Lipid-Droplets. Ein Charakteristikum der Δldh1-Mutante ist das Auftreten übergroßer Lipid-Droplets sowie einer übermäßigen Akkumulation nicht-polarer Lipide und Phospholipiden nach Wachstum auf Medium mit Ölsäure als alleinige Kohlenstoffquelle. Basierend auf den Daten wird eine Funktion von Ldh1p in der Aufrechterhaltung der Lipid-Homeostase in der Hefe durch die Regulation des Spiegels an Phospholipiden wie auch nicht-polaren Lipiden diskutiert. Der peroxisomale Matrix Proteinimport wird durch zyklisierende Rezeptoren ermöglicht, die zwischen dem Zytosol und der peroxisomalen Membran pendeln. Die Ubiquitinilierung des Rezeptors dient dabei als dessen Exportsignal. Ein entscheidender Schritt innerhalb dieses Zyklus ist die ATP-abhängige Ablösung des Rezeptors von der peroxisomalen Membran. Dieser Schritt wird durch die peroxisomalen AAA ATPasen Pex1p und Pex6p bewerkstelligt. In der vorliegenden Arbeit konnte gezeigt werden, dass der AAA-Komplex sowohl die Pex5p-Dislokaseaktivität wie auch eine deubiquitinilierende Aktivität beinhaltet. Im Einklang mit dieser Beobachtung konnte Ubp15p, eine Ubiquitin-Hydrolase, als neuer Bestandteil des AAA-Komplexes identifiziert werden. Ubp15p ist partiell peroxisomal lokalisiert und in der Lage Ubiquitinreste vom modifizierten PTS1-Rezeptor Pex5p abzuspalten. Des Weiteren weisen Ubp15p-defiziente Zellen einen stress-induzierten PTS1-Importdefekt auf. Diese Ergebnisse führen zu dem Modell nachdem die Entfernung des Ubiquitins von Pex5p ein spezifisches Ereignis darstellt welches ein wesendlicher Schritt im RezeptorRecycling darstellt. ABKÜRZUNGSVERZEICHNIS AAA ATP ATPases Associated with various cellular Activities Adenosine triphosphate BPC 1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-sindacene-3undecanoyl)-sn-glycero-3-phosphocholine (bis-BODIPY-FL C11-PC) Cell division cycle 48 protein Candida rugosa lipase 1,2-O-dilauryl-rac-glycero-3-glutaric acid (6-methyl resorufin) ester 1,2-dioleoyl-3-(pyren-1-yl) decanoyl-rac-glycerol Cdc48p CRL DGR DPG DUB ERAD GFP Deubiquitinating enzyme Green Fluorescence Protein E1 E2 E3 Ubiquitin-activating enzymes Ubiquitin-conjugating enzymes Ubiquitin ligases E. coli min ml NPL NSF Escherichia coli Minutes Millilitre Non-polar lipids N-ethylmaleimide sensitive factor (fusion protein) NTD N-terminal domain pex PC PE PL PLA PLC PLD PMPs peroxisome assembly Phosphatidylcholine Phosphatidylethanolamine Polar lipids Phospholipase A Phospholipase C Phospholipase D Endoplasmic-Reticulum-Associated protein Degradation PNB Peroxisome Membrane Proteins p-Nitrophenyl butyrate PNS ProtA PTS RING post nuclear supernatant Protein A Peroxisomal Targeting Signal really interesting new gene SRH Second region of homology TEV Ub Tobacco Etch virus Ubiquitin Ubc VCP Ubiquitin-conjugating enzyme Valosin-containing protein Characterization of peroxisome- and lipid droplet-related proteins of Saccharomyces cerevisiae Dissertation to obtain the degree Doctor Philosophiae (Doctor of Philosophy, PhD) at the Faculty of Biology and Biotechnology Ruhr-University Bochum International Graduate School of Biosciences Ruhr-University Bochum Department of Systems Biochemistry submitted by Dipl.-Biol. & Biochem. Mykhaylo O. Debelyy from Dnepropetrovsk, Ukraine Bochum July, 2011 ERKLÄRUNG Hiermit erkläre ich, dass ich die Arbeit selbständig verfasst und bei keiner anderen Fakultät eingereicht und dass ich keine anderen als die angegeben Hilfsmittel verwendet habe. Es handelt sich bei der heute von mir eingereichten Dissertation um sechs in Wort und Bild völlig übereinstimmende Exemplare. Weiterhin erkläre ich, dass digitale Abbildung nur die originalen Daten enthalten und in keinem Fall inhaltsverändernde Bildbearbeitung vorgenommen wurde. Bochum, den _______________________________________ (Unterschrift) Vorsitzender der Prüfungskommission: Prof. Dr. Franz Narberhaus (Fakultät für Biologie, RUB) 1. Gutachter: Prof. Dr. Ralf Erdmann (Medizinische Fakultät, RUB) 2. Gutachter: Prof. Dr. Ulrich Kück (Fakultät für Biologie, RUB) 3. Gutachter: PD Dr. Mathias Lübben (Fakultät für Biologie, RUB) INDEX__________________________________________________________________ INDEX 3 CHAPTER 1. INTRODUCTION ABSTRACT 1.1 Biology of peroxisomes 1.1.1 Structure and function of peroxisomes 1.1.2 Biogenesis of peroxisomes 1.1.3 Posttranslational modifications of peroxins 1.1.4 Peroxisomal AAA-ATPase peroxins 1.2 Biology of lipid droplets 1.2.1 Structure and function of lipid droplets 1.2.2 Biogenesis of lipid droplets 1.2.3 Interactions between peroxisome and lipid droplets 1.3 Objectives 4 5 5 5 7 12 14 19 19 21 22 24 CHAPTER 2. ORIGINAL WORKS 2.1 Biology of peroxisomes 2.1.1 Lpx1p is a peroxisomal lipase required for normal peroxisomes morphology 2.1.2 The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p 2.1.3 Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery 2.2 Biology of lipid droplets 2.2.1 The putative Saccharomyces cerevisiae hydrolase Ldh1p is localized to lipid droplets 2.2.2 Involvement of the Saccharomyces cerevisiae hydrolase Ldh1p in lipid homeostasis 25 25 CHAPTER 3. DISCUSSION 3.1 Novel hydrolases of S. cerevisiae 3.2 Ubp15p, a novel compound of AAA-complex 77 77 83 CHAPTER 4. REFERENCES 90 CHAPTER 5. MISCELLANEOUS 5.1 Publications 5.2 Personal contribution to the papers 5.3 Conferences 5.4 Curriculum Vitae 5.5 Acknowledgement 5.6 Global scientific outlook for human race 104 104 105 106 107 108 109 25 36 42 65 65 71 3 CHAPTER 1. INTRODUCTION_____________________________________________ 4 ABSTRACT The peroxisomal and lipid droplets related proteins of yeast S. cerevisiae were characterized in this work. Lpx1p and Ldh1p are putative hydrolases and/or lipases of peroxisome and lipid droplets respectively; Pex1p and Pex6p are peroxisomal AAA ATPases; and Ubp15p is a deubiquitinating enzyme. It was shown that Lpx1p is present in the peroxisome but Ldh1p is predominantly localized to lipid droplets. Lpx1p as well as Ldh1p comprises the typical GXSXG-type lipase motif of members of the α/β-hydrolase family. Both proteins carry a putative peroxisomal targeting signal type-1 (PTS1) and can be aligned with two regions of homology. While Lpx1p was shown to be a peroxisomal enzyme, subcellular localization studies revealed that Ldh1p is predominantly localized to lipid droplets. It was shown that Lpx1p import is dependent on the PTS1 receptor Pex5p. Moreover, it was shown that Lpx1p is piggyback-transported into peroxisomes. But it was demonstrated that targeting of Ldh1p to lipid droplets occurs independently of the PTS1 receptor Pex5p. Triacylglycerol lipase as well as hydrolase activities were shown for both recombinant proteins Lpx1p and Ldh1p in vitro. It was shown that the Lpx1p protein is not required for wild-type-like steady-state function of peroxisomes, which might be indicative of a metabolic rather than a biogenetic role. It was clearly shown that peroxisomes in Δlpx1 mutants have an aberrant morphology characterized by intraperoxisomal vesicles or invaginations. It was shown that Ldh1p is not required for the function and biogenesis of peroxisomes. Ldh1p is required for the maintenance of a steady-state level of the nonpolar and polar lipids of lipid droplets. A characteristic feature of the Δldh1 strain is the appearance of giant lipid droplets and an excessive accumulation of nonpolar lipids and phospholipids upon growth on medium containing oleic acid as a sole carbon source. Ldh1p is thought to play a role in maintaining the lipid homeostasis in yeast by regulating both phospholipid and nonpolar lipid levels. It is known that the peroxisomal matrix protein import is facilitated by cycling receptors shuttling between the cytosol and the peroxisomal membrane. One crucial step in this cycle is the ATP-dependent release of the receptors from the peroxisomal membrane. This step is facilitated by the peroxisomal AAA ATPases Pex1p and Pex6p with ubiquitination of the receptor being the main signal for its export. It was shown in this work that the AAA-complex contains Pex5p dislocase as well as deubiquitinating activity. Ubp15p, an ubiquitin hydrolase, was identified as novel constituent of the complex. Ubp15p partially localizes to peroxisomes and is capable to cleave off ubiquitin-moieties from the PTS1-receptor Pex5p. Furthermore, Ubp15p-deficient cells are characterized by a stress related PTS1-import defect. The results merge to a picture in which removal of ubiquitin of the PTS1-receptor Pex5p is a specific event and might represent a vital step in receptor recycling. CHAPTER 1. INTRODUCTION_____________________________________________ 5 CHAPTER 1. INTRODUCTION 1.1 Biology of peroxisomes 1.1.1 Structure and function of peroxisomes Peroxisomes or microbodies are a class of structurally and functionally related ubiquitous eukaryotic organelles that are involved in lipid and antioxidant metabolism (179). Originally these structures were described as cellular organelles in 1966 by C. de Duve and P. Baudhuin (33) after they had been first mentioned in a PhD thesis of J. Rhodin a more than a decade earlier (177). Generally, peroxisomes are spherical organelles with diameter from 0.1 to 1 µm envelop by a single phospholipid bilayer membrane (218) (Fig. 1.1.1.1). Fig. 1.1.1.1 Induction of peroxisomes in yeast S. cerevisiae by oleic acid. Localization and morphology of peroxisome. (A) Red staining: labeling of peroxisome by DsRed-PTS2; Green staining: partial labeling of peroxisome by GFP-Ubp15p; (B) Electron microscopy image of wild-type; (C) Electron microscopy image of peroxisome free mutant (Δpex19); C, cytosol; ER, endoplasmic reticulum; L, lipid droplets; M, mitochondria; N, nucleus; P, peroxisome. The peroxisome-family includes peroxisomes, glyoxysomes of plants and fungi, glycosomes of trypanosomes, and Woronin-bodies of filamentous fungi (11, 138, 179) (Fig.1.1.1.2). The unique variability in function of peroxisomes is displayed by an electron-dense proteinaceous organellar matrix that contains no DNA (179), but is extremely variable in their enzyme content, adjusted to metabolic functions according to the cellular needs. CHAPTER 1. INTRODUCTION_____________________________________________ 6 Fig. 1.1.1.2 Induction of peroxisomes in yeast species and filamentous fungi. Peroxisomes can have highly variable sizes and shapes. Furthermore, they can be present in clusters but can also be dispersed throughout the cytoplasm. A) Aspergillus tamarii cell grown on oleate showing peroxisomes. In addition, Woronin bodies are present near the septum (arrow). Many lipid bodies are present. B) Hansenula polymorpha cell from a methanol-limited chemostat. More than 80% of the cell is filled with cuboid-shaped peroxisomes. C) Saccharomyces cerevisiae cell grown on oleate showing clustered peroxisomes. D) Penicillium chrysogenum hyphae producing the fluorescent peroxisomal protein green fluorescent protein-Ser-Lys-LeuCOOH (GFP-SKL). Cells were grown in penicillin-producing medium and treated with Mitotracker Orange to stain the mitochondria. The bar represents 1 mm, unless indicated otherwise. L, lipid body; M, mitochondrion; N, nucleus; P, peroxisome; V, vacuole. Taken with modifications from (107). The peroxisomal matrix harbours at least 50 different enzymes that are linked to diverse biochemical pathways (96). The β-oxidation of fatty acids and the detoxification of hydrogen peroxide are regarded as the central function of peroxisomes (191, 219). But there is one exception: the Woronin-bodies, function of which is only the plugging of the septal pores in case of hyphal injury. While the ß-oxidation in fungi and plants exclusively take place in peroxisomes (179, 191, 219), in mammalian cells only the very long chain fatty acids are oxidized in peroxisomes (97, 121). Moreover, peroxisomes are involved in the synthesis of CHAPTER 1. INTRODUCTION_____________________________________________ 7 plasmalogens (which contribute more than 80% of the phospholipid content of the white matter in the brain), cholesterol and bile acids (17, 76, 118, 119) as well as the oxidation of alcohols, metabolism of prostaglandins, catabolism of purines and polyamines, the main reaction of photorespiration in plant leafs and final steps of penicillin biosynthesis in some filamentous fungi (80, 153, 215, 216, 218). They are the source of signalling molecules such as jasmonates in plants (36, 160, 225) or lipid-derived ligands for PPARs (peroxisomeproliferator-activated receptors) in humans (51). The existence of severe inherited diseases in human caused by malfunctions in peroxins, encoded by PEX genes, stimulates intensive research interest to the field of peroxisome biogenesis. So far 34 peroxins were discovered to be involved in different stages of peroxisome biogenesis (217). Human peroxisomal disorders can be categorized as either single-enzyme disorders or peroxisomal biogenetic defects (229). Single-enzyme disorders, such as for example Refsum disease is caused by a defect of phytanoyl-CoA hydroxylase, whereas X-linked adrenoleukodystrophy is caused by a defect in a peroxisomal ATPtransporter. In contrast, biogenesis defects are mostly caused by mutations in the PEX genes (211). Peroxisomal disorders are associated with morphological peroxisomal defects such as inclusions or invaginations (56, 151). 1.1 Biology of peroxisomes 1.1.2 Biogenesis of peroxisomes As peroxisomes do not contain genetic material, their protein content is determined by the import of nuclear encoded proteins (26). Peroxisomes can multiply by division (152) or de novo by budding from the endoplasmic reticulum (84, 123). Without exception, peroxisomal matrix proteins are synthesized on free ribosomes and are subsequently imported in a posttranslational manner (136, 188). Like the sorting of proteins to other cellular compartments, protein targeting to peroxisomes depends on signal sequences. Peroxisomal import of most matrix proteins depends on the conserved PTS1 (peroxisomal targeting signal type 1) receptor Pex5p, which recognizes the PTS1 localized at the very C-terminus of the cargo proteins (170, 212). The three-amino-acid signal SKL (serine–lysine–leucine) was the first PTS1 to be discovered, and is in many cases sufficient for directing a protein to peroxisomes. Based on mutagenesis experiments, amino acid permutations and sequence comparisons between different species, the PTS1 generally fits the consensus (S/A/C)-(K/R/H)-(L/M). A second peroxisomal targeting signal (PTS2) (188) is present in considerably fewer peroxisomal CHAPTER 1. INTRODUCTION_____________________________________________ 8 proteins. PTS2 is usually located within the first 20 amino acids of the protein, and has been defined as (RK)-(LVIQ)-XX-(LVIHQ)-(LSGAK)-X-(HQ)-(LAF) (166). PTS2-bearing proteins are recognized by the cytosolic conserved receptor Pex7p (188). Based on the concept of cycling receptors (38, 144), the matrix protein import can be divided into four steps: 1) receptor-cargo recognition in the cytosol, 2) docking at the peroxisomal membrane, 3) cargo-translocation and release, and 4) receptor release from the membrane and recycling. After the cargo recognition by their cognate receptor in the cytosol (72), in yeast the second step in receptor cycle is facilitated by Pex14p together with Pex13p and Pex17p form the docking subcomplex at the peroxisomal membrane and interact in this cycle with both soluble import receptors Pex5p and Pex7p (176) (Fig. 1.1.2.1). Fig. 1.1.2.1 The receptor cycle. According to the model of the cycling receptor, the peroxisomal protein import conceptually can be divided in five steps: (I) cargo recognition in the cytosol and (II) docking of the receptor–cargo complexes to the peroxisomal membrane. (III) Cargo-translocation into the peroxisomal matrix. (IV) Disassembly of the receptor–cargo complex and (V) export of the receptor back to the cytosol. PTS1-containing proteins are recognized by the soluble import receptor Pex5p in the cytosol. Proteins harbouring the PTS2 are recognized by Pex7p and the cofactors Pex18p and Pex21p in S. cerevisiae, the orthologous Pex20p in other fungi or Pex5L in plants and mammals. After this step, the receptor–cargo complex targets to and associates with the peroxisomal membrane via the docking complex consisting of Pex14p, Pex13p and Pex17p. The transport of PTS1-proteins across the membrane is facilitated by formation of a pore mainly consisting of Pex14p and Pex5p. Pex8p connects the RING-complex to the docking complex. The three ubiquitin ligases Pex2p, Pex10p and Pex12p form the RING-complex and together with ubiquitin conjugating enzymes like Pex4p are responsible for receptor ubiquitination. In the last step of the cycle, the receptor Pex5p is exported back to the cytosol by the two AAA-peroxins Pex1p and Pex6p and is enabled for the next round of import. Taken from (179). CHAPTER 1. INTRODUCTION_____________________________________________ 9 It was demonstrated by using the yeast two-hybrid system and pull-down assays that yeast S. cerevisiae Pex5p directly interacts with two separate regions of Pex14p, amino acid residues 1-58 and 235-308. The latter binding site at the C-terminus of Pex14p overlaps with a binding site of Pex7p at amino acid residues 235-325 (158). The functional assessment of these two binding sites of Pex14p with the PTS-receptors indicates that they have distinct roles. Deletion of the N-terminal 58 amino acids caused a partial defect of matrix protein import in Δpex14 cells expressing the Pex14-(59-341)-p fragment; however, it did not lead to a pex phenotype. In contrast, truncation of the C-terminal 106 amino acids of Pex14p completely blocked this process (158). It was proposed that the C-terminus of Pex14p contains the actual docking site and that the N-terminus could be involved in a Pex5p-Pex14p association inside the peroxisomal membrane (158). The molecular mechanism of how the cargo proteins traverse the peroxisomal membrane remains unclear. However, recent reports demonstrated the transient formation of a dynamic pore which is adapted to the size of the cargo and could facilitate the translocation of at least 9 nm particles (222). The final step in the receptor cycle is the release of the receptor back to the cytsosol to facilitate a new round of import. With respect to the PTS1-receptor Pex5p, recent reports demonstrated that its dislocation from the peroxisomal membrane to the cytosol at the end of the receptor cycle is ATP-dependent and catalyzed by the AAA-peroxins Pex1p and Pex6p (150, 172). With this respect in accordance to the export-driven import model it is believed, that the export of receptor delivers the energy for cargo-translocation (184). Pex4p-catalysed mono-ubiquitination of Pex5p direct the receptor for recycling, thereby enabling further rounds of matrix protein import, whereas Ubc4p-catalysed polyubiquitination targets Pex5p to proteasomal degradation (44, 106, 171, 172). The import of peroxisomal membrane proteins (PMPs) is differing from the import machinery of peroxisomal matrix proteins (43, 68). This is in agreement with the fact that most pex-mutants are distinguished by an affected import of matrix proteins but not affected import of PMPs. In these mutants, the PMPs are imported in peroxisomal remnants, so called ghosts (25, 183, 188). Only several mutants were characterized by the complete absence of detectable peroxisomal membrane ghosts. Functional complementation of these mutants led to the identification of Pex3p, Pex19p and in some organisms Pex16p which are involved in the biogenesis of the peroxisomal membrane (10, 41, 60, 67, 88, 146, 179, 200) (Fig. 1.1.2.2). CHAPTER 1. INTRODUCTION_____________________________________________ 10 Fig. 1.1.2.2 Topogenesis of peroxisomal membrane proteins. Two routes are proposed for the targeting of peroxisomal membrane proteins (PMPs). Class I proteins are directly imported into existing peroxisomes. Class II proteins are first targeted to ER where they concentrate in pre-peroxisomal vesicles which then are targeted to existing peroxisomes or function as an origin for de novo formation of peroxisomes. Currently, it is controversially discussed whether class I PMPs are also targeted to the ER and whether class II PMPs are also targeted to existing peroxisomes. Taken from (179). Various roles have been suggested for Pex19p. Initially, due to its capacity to interact with the majority of the peroxisomal membrane proteins (PMPs), and according, to its multiple localization at the peroxisomal membrane and in the cytosol, Pex19p is considered to be a soluble import receptor for newly synthesized PMPs (100, 185). As a consequence, Pex19p binds PMPs in the cytosol and delivers them to the peroxisomal membrane by docking to its membrane anchored binding partner Pex3p (67, 179) (Fig. 1.1.2.3). Subsequent, Pex19p is additionally assumed to behave as a PMP-specific chaperone. Correspondingly, Pex19p bear the capacity to adhere and sustain PMP by the development of a soluble complex and in this manner anticipating conglomeration of the PMP (105, 192). Also, Pex19p has possibility to function as an insertion factor during PMP import (54, 198) or act as an assembly/disassembly factor for peroxisomal membrane complexes at the peroxisomal membrane (52, 179). Recently, it was shown that Pex19p is required for the transport of Pex3p from the endoplasmic reticulum (ER) to the peroxisomal membrane (84). Pex3p is an integral membrane protein at the peroxisomal membrane with a topology distinct all over species (60, 75, 93, 179, 199). CHAPTER 1. INTRODUCTION_____________________________________________ 11 Fig. 1.1.2.4 Pex19p-dependent import of PMPs. Class I peroxisomal membrane proteins (PMPs) harbour a peroxisomal membrane protein targeting signal (mPTS) which is recognized in the cytosol by the import receptor and/or PMP-specific chaperone Pex19p, a farnesylated, mostly cytosolic protein with a small portion of the protein found associated with the peroxisomal membrane. In the next step, the cargo-loaded Pex19p docks to the peroxisomal membrane via association with its docking factor Pex3p. Then the PMP is inserted into the membrane in an unknown manner but presumably with assistance of Pex19p, Pex3p and, in some organisms, Pex16p. The requirement of ATP for this process is not clear. Finally, Pex19p shuttles back to the cytosol where it might start a new round of import. Taken from (179). In Saccharomyces cerevisiae, Pex3p bear an N-terminal transmembrane region and a large Cterminal domain to be turned toward the cytosolic side of the peroxisome (86, 179). Pex3p performs a pivotal function in the import of PMPs at which point it assists as a docking factor at the peroxisomal membrane and acts as binding partner for Pex19p-PMP-complexes meanwhile import of the PMPs (49, 53, 147). Pex3p additionally acts as a crucial factor in the de novo development of peroxisomes as it is considered to be the initiating step for this peroxisome assembling action. PMP insertion into the peroxisomal membrane in some organism required assistance of Pex16p. It was demonstrated that Pex16p is an integral membrane protein which is mainly found in higher eukaryotes (129, 179, 200) and in the yeast Y. lipolytica (41). Although the mammalian Pex16p is an integral membrane protein with the C- as well as the N-terminus facing the cytosol (87), the yeast Pex16p is a membrane associated protein facing the peroxisomal lumen (41). It was shown that Pex16p execute distant activities in peroxisome CHAPTER 1. INTRODUCTION_____________________________________________ 12 biogenesis. The mammalian Pex16p is required for the topogenesis of membrane proteins and acts in the very early stages of peroxisome biogenesis while the yeast Pex16p is a negative regulator of peroxisomal fission (41, 108, 179). 1.1 Biology of peroxisomes 1.1.3 Posttranslational modifications of peroxins The theory of cycling receptors Pex5p and Pex7p imply consecutive interaction of the receptors to distinct proteins or protein complexes at the peroxisome (116, 158). The regulation of such interactions are implementing by reversible posttranslational modification such as phosphorylation and/or ubiquitination (116, 171). Actually, the membrane proteins Pex14p and Pex15p were shown to be phosphorylated (42, 99, 114). Nevertheless, the physiological functions of phosphorylation in peroxisomal matrix protein import are unexplored (116). Recently, two peroxins have been shown to be ubiquitinated. For example, Pex18p, a protein involved in the PTS2 pathway, is constitutively degraded in an ubiquitin-dependent manner (174). Considering such observation two independent research groups demonstrated polyubiquitination of Pex5p in cells deficient in constituents of the AAA or Pex4p-Pex22p complexes (106, 171). Besides, it was shown for Pex5p that polyubiquitination leads to the proteasomal degradation (171). It was demonstrated that ubiquitination of proteins requires the consecutive activity of at least three types of enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (167, 226) (Fig. 1.1.3.1). At the terminating stage of the ubiquitination cascade an isopeptide bond amongst ubiquitin and the lysine residue of the substrate is arranged. This reaction is catalyzed by the E2 enzyme, usually in association with the E3 ligase. The length of the ubiquitin chain conjugated to a protein substrate is carrying out a considerably meaningful function. Polyubiquitinated proteins (the minimal chain length is four ubiquitin moieties) are normally distinguished from other non-ubiquitinated proteins and degraded by the proteasome (213). In contrast, monoubiquitination, an attachment of a single ubiquitin moiety, regulates cellular processes such as endocytosis, sorting into multivesicular bodies and virus budding in a proteasome-independent way (81). It was shown that Pex5p is a monoubiquitinated and stable protein in wild-type cells. Pex5p monoubiquitination occurs at the peroxisome and is blocked in cells deficient of operative docking or RING finger complexes, evoking concept that Pex5p CHAPTER 1. INTRODUCTION_____________________________________________ 13 monoubiquitination is a late event in peroxisomal matrix protein import (116, 171). It was shown that polyubiquitination of Pex5p protein is not a prerequisite for functional peroxisomal protein import in S. cerevisiae (169). Fig. 1.1.3.1 The ubiquitylation pathway. Free ubiquitin (Ub) is activated in an ATPdependent manner with the formation of a thiol-ester linkage between E1 and the carboxyl terminus of ubiquitin. Ubiquitin is transferred to one of a number of different E2s. E2s associate with E3s, which might or might not have substrate already bound. For HECT domain E3s, ubiquitin is next transferred to the active-site cysteine of the HECT domain followed by transfer to substrate (S) (as shown) or to a substrate-bound multi-ubiquitin chain. For RING E3s, current evidence indicates that ubiquitin might be transferred directly from the E2 to the substrate. Taken with modifications from (226). It was shown that Pex5p is a monoubiquitinated and stable protein in wild-type cells. Pex5p monoubiquitination occurs at the peroxisome and is blocked in cells deficient of operative docking or RING finger complexes, evoking concept that Pex5p monoubiquitination is a late event in peroxisomal matrix protein import (116, 171). It was shown that polyubiquitination of Pex5p protein is not a prerequisite for functional peroxisomal protein import in S. cerevisiae (169). Moreover, it was shown that polyubiquitinated forms of Pex5p concentrate in definite pex mutants in an Ubc4p-dependent fashion (116), an observation that is in agreement with previous reports (106, 171). Despite, monoubiquitination of Pex5p in wild-type cells is not controlled by Ubc4p, and peroxisome biogenesis is not disturbed in cells deficient of Ubc4p (116). Besides, it was demonstrated the polyubiquitination of Pex5p is part of a quality control system that direct membraneaccumulated Pex5p for proteasomal degradation (44, 106, 172). The protein Pex19p acts as a receptor and chaperone of peroxisomal membrane proteins (PMPs) (190). The conserved CaaX box peroxin Pex19p is known to be modified by CHAPTER 1. INTRODUCTION_____________________________________________ 14 farnesylation (67, 104, 146). It was recently shown that the complete pool of Pex19p is processed by farnesyltransferase in vivo and that this modification is independent of peroxisome induction or the Pex19p membrane anchor Pex3p. Moreover, it was demonstrated that genomic mutations of PEX19, which blocks farnesylation are critical for correct matrix protein import into peroxisomes. It was shown that mutants defective in Pex19p farnesylation are characterized by a significantly reduced steady-state concentration of prominent peroxisomal membrane proteins Pex11p and Ant1p as well as constitutive compounds of the peroxisomal import machinery such as RING peroxins (180). 1.1 Biology of peroxisomes 1.1.4 Peroxisomal AAA-peroxins The highly diverse and adaptive character of peroxisomes is accomplished by modulation of their enzyme content, which is mediated by dynamically operating protein-import machineries. The import of matrix proteins into the peroxisomal lumen has been described as the ATP-consuming step. It was shown that the peroxisomal AAA-ATPase (ATPase Associated with various cellular Activities) proteins Pex1p and Pex6p are mechano-enzymes and core components of a complex which dislocates the cycling import PTS1-receptor Pex5p from the peroxisomal membrane back to the cytosol. Such release of Pex5p has been regarded as the final step of the peroxisomal protein import cascade. The AAA-mediated process is regulated by the ubiquitination status of the receptor Pex5p. Pex4p-catalysed monoubiquitination of Pex5p primes the receptor for recycling, thereby enabling further rounds of matrix protein import, whereas Ubc4p-catalysed polyubiquitination targets Pex5p to proteasomal degradation (168). AAA-proteins are characterized by a typical modular architecture as they contain an N-terminal non-ATPase domain which is followed by at least one conserved AAA domain. Each AAA-cassette usually contains an ATP-binding site (Walker A) and an ATP-hydrolysis site (Walker B) along with other motifs, such as the SRH (46) (Fig 1.1.4.1). Pex1p and Pex6p are type II AAA-proteins, which are characterized by two AAA domains. In both AAA peroxins, the second AAA domain is more conserved than the first one. Interaction and subsequent oligomerization of Pex1p and Pex6p is believed to be initiated in the cytosol and involves their first less conserved AAA domains (D1) (19, 204). Although neither binding nor hydrolysis of ATP at D1 seems to be essential for functionality in both yeast and humans, the interaction of human Pex1p and Pex6p is CHAPTER 1. INTRODUCTION_____________________________________________ 15 stimulated by binding of ATP to D1 of human Pex1p and Pex6p (48, 204). Furthermore, ATP binding, but not hydrolysis, at the second AAA cassette (D2) of Pex1p is required for the Pex1p–Pex6p interaction in both systems (19, 204). Fig 1.1.4.1 Molecular organization of the AAA complex in S. cerevisiae. The AAA peroxins Pex1p and Pex6p are composed of an NTD, a non-conserved AAA domain (D1) and a conserved AAA domain (D2). The AAA domains contain ATP-binding sites (A) and, with the exception of D1 of Pex6p, also ATP-hydrolysis sites (B). Pex1p and Pex6p form a heteromeric complex, and oligomerization requires the presence of the D1 domains and is stimulated by ATP binding to Pex1p D2. Recruitment of the AAA complex to peroxisomes occurs via binding of Pex6p NTD to Pex15p and requires ATP binding at Pex6p D1, while detachment from Pex15p needs ATP binding and hydrolysis at Pex6p D2. The peroxisomal AAA complex dynamically associates with the functional matrix protein-import machinery (importomer) and Pex4p (Ubc10p) is supposed to be required for the disconnection of the AAA complex from the importomer. Taken with modifications from (168). Pex1p and Pex6p are believed to form heterohexameric structures in the cytosol and at the peroxisomal membrane (47, 178, 203, 204). However, it is not clear whether formation of a heteromeric assembly of the AAA peroxins is a prerequisite for their function, as one population of Pex1p does not co-localize with Pex6p in mammalian cells (204, 228). Although the formation of hexameric structures is common to AAA proteins, the formation of heterohexamers has been found in few other cases, such as the m-AAA (matrix CHAPTER 1. INTRODUCTION_____________________________________________ 16 AAA) complex, consisting of Yta10p and Yta12p, which is active at the matrix site of the inner mitochondrial membrane, (7) or the six different Rpt ATPases from the 19S proteasome (62). The recruitment of AAA-complexes to peroxisomes is mediated by the tail-anchored peroxisomal membrane proteins Pex15p in S. cerevisiae or its functional orthologue Pex26p in human cells via binding of the N-terminal domain of Pex6p, stimulated by ATP binding to the Walker A motif of Pex6p D1 (20, 145). In contrast, the Walker A and B motifs of Pex6p D2 are required for an efficient detachment from Pex15p/Pex26p (20, 57, 204). Although Pex15p and Pex26p have been described as adaptor proteins for the N-terminal part of Pex6p, no adaptor has yet been identified for Pex1p (Fig. 1.1.4.2). Fig. 1.1.4.2 Schematic representation of interaction between Pex15p and Pex6p. The Nterminus of Pex15p interacts with the N-terminal part of Pex6p, an interaction which is stimulated by ATP-binding to the first AAA domain (A1) of Pex6p. On the other side hydrolysis of ATP by the second AAA domain of Pex6p (B2) stimulates release of Pex6p from Pex15p. Taken from (20). The NTD (N-terminal domain) of murine Pex1p represents the only available crystal structure of the AAA peroxins (193). The NTD folds into two structurally independent globular subdomains (N- and C-lobe), which comprise an N-terminal double-ψ fold and a Cterminal β-barrel, separated by a shallow groove. Similar grooves were found in the adaptorbinding sites within the NTDs of VCP, NSF and VAT (VCP-like ATPase from Thermoplasma), suggesting functional similarity (193). The Pex15p-anchored AAA complex itself is part of an even larger protein complex at the peroxisomal membrane, the peroxisomal matrix protein import machinery called the CHAPTER 1. INTRODUCTION_____________________________________________ 17 importomer (178). To conclude, at least in S. cerevisiae, the Pex1p-bound nucleotides seem to influence the Pex1p–Pex6p interaction, while the different nucleotide states of Pex6p regulate the dynamic Pex6p–Pex15p/Pex26p association. The non-conserved domains are responsible for oligomerization, while the conserved domains exhibit the main ATPase activity. Import of folded proteins into peroxisomes occurs in a post-translational manner and depends on ATP. The soluble PTS1 receptor Pex5p is the major signal-recognition factor of proteins destined for the peroxisomal matrix. The receptor cycle of Pex5p involves cargo recognition in the cytosol, docking of the receptor–cargo complex to the peroxisomal membrane, translocation of the receptor–cargo complex to the luminal side of the membrane, followed by release of the cargo into the matrix and retrotranslocation of the receptor back to the cytosol (44). Permeabilized cell systems of human fibroblasts provided the first evidence that Pex5p accumulated reversibly at the peroxisomal membrane under ATP-modulated conditions (38). Detailed in vitro studies revealed that the binding and translocation of Pex5p itself is ATPindependent while the export of Pex5p back to the cytosol requires ATP (69). The identity of the corresponding ATPase remained a matter of debate until in vitro systems in S. cerevisiae (172) and human fibroblast cells (150) identified Pex1p and Pex6p as the motor proteins of Pex5p export. Their function in this process requires the presence of their membrane-anchor proteins, Pex15p or Pex26p. The in vitro reconstitution of the complete Pex5p cycle revealed that ATP binding and hydrolysis at both Pex1p D2 and Pex6p D2 is needed for receptor dislocation (172). Interestingly, the Walker B motif of Pex1p D2 seems to have no function in formation or targeting of the AAA complexes (19, 204) and thus may be exclusively required for handling of the substrate. The binding and consumption of ATP is believed to induce conformational changes within the AAA peroxins that generate the driving force to pull the receptor out of the membrane by a mechanism possibly similar to the one of Cdc48p (p97/VCP) in ERAD The mechanism of substrate recognition by the AAA peroxins is not understood. Although Pex5p and the AAA-proteins form a complex at the peroxisomal membrane (150, 172, 178), no direct interaction of the PTS1 receptor with either Pex1p or Pex6p has been reported. This interaction seems to be regulated or mediated by a third factor, which could represent an unknown adaptor protein of the AAA-peroxins or post-translational modification of the substrate. It is well known that both processes play a central role in the function of Cdc48p (p97/VCP) (98, 227), which is the closest evolutionary relative of Pex1p and Pex6p (58, 186). As a consequence, the question has to be addressed of how the AAA-peroxins can CHAPTER 1. INTRODUCTION_____________________________________________ 18 distinguish Pex5p forms destined for dislocation from cargo-loaded Pex5p species destined for cargo translocation. A possible solution may arise from the crystal structure of Pex1p NTD, which displays similarities to the corresponding adaptor-binding domains of other AAA proteins (193). Data from p97 and Ufd1 have identified a double-ψ β-barrel fold as a ubiquitin-binding domain with binding sites for both mono- and poly-ubiquitin (163). Most interestingly, the PTS receptors Pex5p, Pex18p and Pex20p have been demonstrated to be ubiquitinated (106, 128, 171, 174). The PTS1 receptor Pex5p of S. cerevisiae is monoubiquitinated in wild-type cells (116), whereas it has been shown to be polyubiquitinated in mutants of the proteasome or cells affected in the AAA and Pex4p–Pex22p complexes of the peroxisomal protein-import machinery (106, 171). Polyubiquitination of Pex5p, requiring the ubiquitin conjugating enzymes Ubc4p and the partly redundant Ubc5p and Ubc1p, takes place exclusively at the peroxisomal membrane and marks the receptor for proteasomal degradation as part of a quality-control system (106, 116, 171). Alternatively, Pex5p is the specific molecular target for mono-ubiquitination by Pex4p (Ubc10p) (169, 232), which is essential for peroxisomal biogenesis (231) and is anchored via Pex22p to the peroxisomal membrane (113). The functional role of ubiquitination in the dislocation process has been elucidated by in vitro export assays, revealing that mono-ubiquitination of Pex5p constitute the export signal under physiological conditions, whereas polyubiquitination seems to provide an export signal for the release of dysfunctional PTS1 receptors from the membrane and proteasomal degradation as part of the quality-control pathway (169). The direct mechanistic influence of this modification on the export reaction remains to be investigated. The AAA peroxins may interact directly or indirectly via putative adaptors with the ubiquitin tag on Pex5p. Alternatively, the attachment of ubiquitin may induce local conformational changes within Pex5p to expose hidden binding sites. This mode of interaction is also discussed for Cdc48p (p97/VCP), which binds ubiquitin via adaptor complexes such as Ufd1/Npl4 and via its N-terminal domain. This domain is capable of recognizing ubiquitin chains and also non-modified segments of its substrates (208, 236). Notably, the AAA complex displays significantly increased association with the importomer in PEX4-deficient cells, indicating that the ATPase cycles of Pex1p and Pex6p are coupled to the mono-ubiquitination-dependent receptor cycle of Pex5p (178). CHAPTER 1. INTRODUCTION_____________________________________________ 19 1.2 Biology of lipid droplets 1.2.1 Structure and function of lipid droplets Lipid droplets (LDs) are remarkable dynamic subcellular organelles of globular shape with a size range from 20 to 100 µm, depending on the cell type (37, 50, 73, 201). LDs are depots of neutral lipids with a complex biology that exist in virtually any kind of cell, ranging from bacteria to yeasts, plants, and higher mammals (15, 55, 73). In many cells, LDs occupy a considerable portion of the cell volume and weight (221). As the major intracellular storage organelles, LDs were first described in the works of R. Altmann and E. B. Wilson in the 19th century (2, 233) (Fig. 1.2.1.1). Fig. 1.2.1.1 Lipid droplets in yeast S. cerevisiae. Localization and morphology of lipid droplets in wild-type yeast strain BY4742. (A) Erg6p-RFP labeled lipid droplets; (B) Oil Red O-stained lipid droplets; (C) Electron microscopy image of oleic acid induced yeast cell; C, cytosol; ER, endoplasmic reticulum; L, lipid droplets; M, mitochondria; N, nucleus; P, peroxisome. In contrast to the vesicular organelles, which have the aqueous content enclosed by a phospholipid bilayer membrane (50, 55), mature LDs have a unique physical structure: they have a neutral lipid core consisting of triacylglycerols (TG) and sterol esters (SE) surrounded by a phospholipid monolayer (15, 132, 239) that contains numerous peripheral or embedded proteins (143, 207) (Fig. 1.2.1.2). CHAPTER 1. INTRODUCTION_____________________________________________ 20 Fig. 1.2.1.2 Lipid droplets composition. Taken with modifications from (73). TG, as well as SE, play a crucial role for the cell: TG is the main energy store, and both TG and SE are depots of membrane lipid components (221). LDs can tightly regulate the level of intracellular free cholesterol by hydrolyzing sterol ester (143). The LD core also contains other endogenous neutral lipids, like monoacylglycerol, diacylglycerol, free cholesterol, and retinol ester, and xenobiotic hydrophobic compounds, such as polycyclic aromatic hydrocarbons (73, 94, 195, 205, 207). A number of proteins are specifically targeted to the LD surface (95), where they can regulate LD dynamics and the turnover of stored lipids (132). Lipid-metabolizing enzymes, including hydrolases and lipases, are the major classes of LD enzymes (37). LDs play crucial roles in cellular energy homeostasis and lipid metabolism (221). LDs can provide a rapidly mobilized lipid source for many important biological processes. Neutral lipids may be mobilized for the generation of energy by β-oxidation (191, 219) or for the synthesis of membrane lipids and signalling molecules (37). It has been shown that all cell types have the ability to generate LDs in response to elevated fatty acid levels and to subsequently metabolize and disperse these LDs when conditions are reversed (143), thereby providing an emergency energy pool for cell survival (15). Due to their unique architecture, LDs can protect cells from the effects of potentially toxic lipid species, such as unesterified lipids (117, 132) or toxic free fatty acids (15), by depositing them inside the LD’s core. In addition to this lipid scavenging function, LDs can transiently store certain proteins, which may be released or degraded at later time points (37, 55, 64, 230). LDs interact with other organelles such as peroxisomes, endosomes, endoplasmatic reticulum (ER), plasma membrane, mitochondria and caveolae (73). The obesity and type 2 diabetes mellitus are most common lipid droplets-associated disorders caused by impairment of triacylglycerol (TAG) metabolism (112). The key anabolic and catabolic enzymes involved in TAG metabolism are conserved between yeast and mammals (8, 32, 175). CHAPTER 1. INTRODUCTION_____________________________________________ 21 1.2 Biology of lipid droplets 1.2.2 Biogenesis of lipid droplets Biogenesis of LDs is tightly connected to the ER (15, 37, 73) (Fig. 1.2.2.1). Several models were recently proposed for description of LDs de novo biosynthesis. In the ERbudding model, the neutral lipids accumulate in the interspace between the bilayer leaflets of the ER membrane that subsequently budding-out of the cytoplasm-oriented phospholipids hemimembrane with formation of the nascent LDs (15, 73). Fig. 1.2.2.1 Lipid droplets in adipocytes. (a) 3T3-L1 adipocytes that have been stimulated to induce lipolysis and then labelled for Rab18 (red) and neutral lipids (green). Rab18 is specifically recruited to the surface of a subset of lipid droplets (LDs). The scale bar represents 10 µm. (b) High-pressure frozen 3T3-L1 adipocytes that were processed for electron-microscopy observation after freeze substitution. Note the complexity of the membranes that wrap around and associate with LDs (such complexity represent result of interaction of lipid droplest with endoplasmic reticulum membranes). The scale bar represents 1 µm. Taken with modifications from (143). In the ER-domain model, the LDs remain fused to the ER and are lipid-bearing protrusions of the ER membrane, developing a specialized ER domain (73). In the bicelle model, neutral lipids aggregate between the two leaflets of the ER membrane but, instead of budding, nascent LDs are excised from the membrane, acquire phospholipids from both the cytosolic and luminal leaflets (73, 173). In the vesicular-budding model, tiny immature bilayer vesicles that remain attached to the ER membrane are exploited as a precursor for LDs formation. Neutral lipids are supplied into the vesicle bilayer and blow up the intermembrane volume, finally squeezing the vesicular lumen so that it becomes a tiny incorporation inside the LDs (73, 221) (Fig. 1.2.2.2). CHAPTER 1. INTRODUCTION_____________________________________________ 22 Fig. 1.2.2.2 Lipid droplets biogenesis models. Taken with modification from (73). 1.2 Biology of lipid droplets 1.2.3 Interactions between peroxisome and lipid droplets Peroxisomes are frequently shown to be tightly associated to lipid droplets (18, 28, 64, 65, 78, 159). It was clearly demonstrated by J. Goodman that lipid droplets and peroxisomes have tight physiological interconnections. It was shown that oleic acid induced peroxisomes of yeast S. cerevisiae are stabily associated with lipid droplets by formation of tubular-shaped protrusions into the lipid droplets cores (18). It was demonstrated that peroxisomes can invade lipid droplets with pexopodia and establish peroxisome – lipid droplets synapses. Such close contacts could facilitate lipid molecules bidirectional transport across two organelles (18). For instance, ether lipids, which are normally synthesized in peroxisomes, were shown to be highly enriched in the lipid droplets core of several cell types (13, 18). Some of the peroxisomes inside of lipid droplets constellations are often shown to be dumbbell-shaped, indicating a dependence of the peroxisomal fission (24, 66) on lipid droplets close physical association (21). It was shown in plants that glyoxysomal (peroxisomal) membrane lipids of germinating cotton seeds have exclusively lipid droplets but not endoplasmic reticulum origin (28). In that case triacylglycerols as well as fatty acids were shown to be directly trafficking from lipid droplets to glyoxysomes. Such an observation can indicate about requirements of lipid droplets in peroxisomal maturation and/or fission (64) in contrary to known fact that endoplasmic reticulum membranes are the source of pre-peroxisomal vesicles (84). Moreover, CHAPTER 1. INTRODUCTION_____________________________________________ 23 it was shown in plant that cotyledons of a Δped1 strain (strains lacking peroxisomal fatty acid β-oxidation pathway) have a substantial portion of tight physical contact of lipid droplets and glyoxysomes, with tubular structures within the glyoxysomes that appear to be derived from lipid droplets; possibly these formations are system of transportation of triacylglycerols for glyoxysomal β-oxidation (78). It was demonstrated that yeast Saccharomyces cerevisiae can form extensive long-term contacts between peroxisome and lipid droplets in case of their culturing on medium containing oleic acid as a sole carbon source; in case of yeast culturing on glucose medium only transient interactions were observed (18). Fungi commonly exhibit peroxisome – lipid droplet intimae association. In yeast Yarrowia lipolytica grown in oleic acid medium, many peroxisomes in a temperature-sensitive Δpex3 mutant strain (strain partially deficient in pre-peroxisome budding from the ER) wrap around lipid droplets, as if attempting to access core lipids for membrane assembly (14, 18). Furthermore, animal cells also were shown to demonstrate an extensive association of peroxisomes with lipid droplets in cultured COS-7 cells (18, 187). CHAPTER 1. INTRODUCTION_____________________________________________ 24 1.3 Objectives The goal of this work was to study the biogenesis of peroxisomes and lipid droplets in yeast S. cerevisiae. In the first part of the thesis, the objectives were to characterize the Lpx1p protein: (1) prove the localization of Lpx1p in the peroxisome; (2) show the role of the peroxisomal targeting signal type 1 (PTS1) in the targeting of Lpx1p to the peroxisome; (3) demonstrate the dependence of Lpx1p transport to the peroxisome on the peroxisomal shuttling receptor Pex5p; (4) prove the existence of piggyback-transport of Lpx1p into the peroxisomes; (5) express the recombinant Lpx1p in Escherichia coli; (6) prove the existence of the hydrolase and triacylglycerol lipase activities for Lpx1p in vitro; (7) investigate the role of Lpx1p in the peroxisome metabolism and/or the biogenesis. In the second part of the thesis, the objectives were to characterize the peroxisomal AAA (ATPases Associated with diverse cellular Activities) ATPase proteins Pex1p and Pex6p. In the third part of the thesis, the objectives were to characterize Ubp15p protein: (1) prove the partial localization of Ubp15p in the peroxisome; (2) express the recombinant Ubp15p in Escherichia coli; (3) prove the existence of the ubiquitin hydrolase, monoUb-Pex5p and/or polyUb-Pex5p deubiquitinating activities for Ubp15p in vitro; (4) demonstrate the role of Ubp15p in peroxisome metabolism and/or biogenesis (show some Ubp15p specific phenotype); (5) show physical interaction of Ubp15p with components of peroxisomal export machinery (Pex6p); (6) prove the existence of Ub-Pex5p dislocase and deubiquitinating activities in the purified from yeast AAA-ATPase complex in vitro (show association of deubiquitinating activity with dislocase activity in the endogenously expressed AAA-ATPase complex); (7) show the role of Ubp15p in the cycle of shuttling receptor Pex5p; (8) prove the requirement of enzymatic deubiquitination of Ub-Pex5p in vivo; (9) show steady state level of Ub-Pex5p in wild-type and Δubp15 yeast strains. In the forth part of the thesis, the objectives were to characterize Ldh1p protein: (1) show the localization of Ldh1p (it was shown localization the lipid droplets); (2) show the role of the peroxisomal targeting signal type 1 (PTS1) in the targeting of Ldh1p to the lipid droplets; (3) demonstrate the dependence of Ldh1p transport to the lipid droplets on the peroxisomal shuttling receptor Pex5p; (4) express the recombinant Ldh1p in Escherichia coli; (5) investigate the existence of the hydrolase and triacylglycerol lipase activities for Ldh1p in vitro; (6) show the role of conserved serine in the hydrolase/lipase motif GXSXG for enzyme activity; (7) elucidate the role of Ldh1p in the lipid droplets metabolism and/or the biogenesis (show some Ldh1p specific phenotype). Lpx1p is a peroxisomal lipase required for normal peroxisome morphology Sven Thoms1,*, Mykhaylo O. Debelyy1, Katja Nau1,†, Helmut E. Meyer2 and Ralf Erdmann1 1 Institut für Physiologische Chemie, Abteilung für Systembiochemie, Ruhr-Universität Bochum, Germany 2 Medizinisches Proteomcenter, Ruhr-Universität Bochum, Germany Keywords lipase; peroxin; peroxisome; proteomics; PTS1 Correspondence R. Erdmann, Abteilung für Systembiochemie, Ruhr-Universität Bochum, Universitätsstr. 150, 44780 Bochum, Germany Fax: +49 234 32 14266 Tel: +49 234 322 4943 E-mail: [email protected] Present address *Universitätsmedizin Göttingen, Abteilung für Pädiatrie und Neuropädiatrie, GeorgAugust-Universität, Germany †Forschungszentrum Karlsruhe, Institut für Toxikologie und Genetik, Germany Lpx1p (systematic name: Yor084wp) is a peroxisomal protein from Saccharomyces cerevisiae with a peroxisomal targeting signal type 1 (PTS1) and a lipase motif. Using mass spectrometry, we have identified Lpx1p as present in peroxisomes, and show that Lpx1p import is dependent on the PTS1 receptor Pex5p. We provide evidence that Lpx1p is piggyback-transported into peroxisomes. We have expressed the Lpx1p protein in Escherichia coli, and show that the enzyme exerts acyl hydrolase and phospholipase A activity in vitro. However, the protein is not required for wild-type-like steadystate function of peroxisomes, which might be indicative of a metabolic rather than a biogenetic role. Interestingly, peroxisomes in deletion mutants of LPX1 have an aberrant morphology characterized by intraperoxisomal vesicles or invaginations. (Received 20 September 2007, revised 22 November 2007, accepted 30 November 2007) doi:10.1111/j.1742-4658.2007.06217.x Peroxisomes are ubiquitous eukaryotic organelles that are involved in lipid and antioxidant metabolism. They are versatile and dynamic organelles engaged in the b-oxidation of long and very long chain fatty acids, in a-oxidation, bile acid and ether lipid synthesis, and in amino acid and purine metabolism [1]. Peroxisomes are a source of reactive oxygen species, and are involved in the synthesis of signalling molecules in plants. Remarkably, peroxisomes are the only site of fatty acid b-oxidation in plants and fungi. Human peroxisomal disorders can be categorized as either single-enzyme disorders or peroxisomal biogenetic defects [2]. Single-enzyme disorders, for example Refsum disease caused by a defect of phytanoyl CoA hydroxylase, or X-linked adrenoleukodystrophy caused by a defect in a peroxisomal ATP-transporter. Biogenetic defects are mostly caused by mutations in the peroxisomal biogenesis genes, the PEX genes, that code for peroxins [3]. Peroxisomal disorders are associated with morphological Abbreviations BPC, 1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-sindacene-3-undecanoyl)-sn-glycero-3-phosphocholine (bis-BODIPY-FL C11-PC); DGR, 1,2-O-dilauryl-rac-glycero-3-glutaric acid (6-methyl resorufin) ester; DPG, 1,2-dioleoyl-3-(pyren-1-yl)decanoyl-rac-glycerol; PNB, p-nitrobutyrate. 504 FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS S. Thoms et al. peroxisomal defects such as inclusions or invaginations [4,5]. Peroxisomal import of most matrix proteins depends on the PTS1 (peroxisomal targeting signal type 1) receptor Pex5p, which recognizes the PTS1 localized at the very C-terminus [6,7]. The three-amino-acid signal SKL (serine–lysine–leucine) was the first PTS1 to be discovered, and is in many cases sufficient for directing a protein to peroxisomes. Most PTS1 are tripeptides of the consensus [SAC][KRH][LM] located at the extreme C-terminus. A second matrix protein peroxisomal targeting signal (PTS2) is present in considerably fewer peroxisomal proteins. PTS2 is usually located within the first 20 amino acids of the protein, and has been defined as [RK][LVIQ]XX[LVIHQ][LSGAK]X[HQ][LAF] [8]. PTS2-bearing proteins are recognized by the cytosolic receptor Pex7p. Systems biology approaches led to the identification of Lpx1p as an oleic acid-inducible peroxisomal matrix protein of unknown function [9,10]. The gene sequence of LPX1 predicts a lipase motif of the GxSxG type that is typical for a ⁄ b hydrolases [11,12]. Using mass spectrometry, we identify Lpx1p as present in peroxisomes, and analyse its peroxisomal targeting. We show that it acts as a phospholipase A, and, by electron microscopy and morphometry, we provide the first evidence for an interesting peroxisomal phenotype of the Dlpx1 deletion mutant. Results Identification of Lpx1p in peroxisomes by mass spectrometry We identified Lpx1p (lipase 1 of peroxisomes; EC 3.1.1.x) in a follow-up study to an exhaustive proteomic characterization of peroxisomal proteins [13]. This involved purification of peroxisomes from oleicacid induced Saccharomyces cerevisiae, and subsequent membrane extraction using low- and high-salt buffers. Low-salt-extractable proteins were solubilized in SDS buffer, and separated by RP-HPLC [14]. Proteins in individual HPLC fractions were further separated by SDS–PAGE, and protein bands were cut out and analysed by mass spectrometry. Lpx1p (systematic name: Yor084wp) was extractable by low salt and identified together with the peroxisomal aspartate aminotransferase Aat2p in HPLC fraction 7 at a molecular mass of approximately 45 kDa (Fig. 1A) [15]. The predicted molecular mass of Lpx1p is 44 kDa. It carries a peroxisomal targeting signal type 1, glutamine–lysine–leucine (QKL) (Fig. 1B,D). The amino Peroxisomal lipase Lpx1p acid sequence comprises the lipase motif GHSMG of the general GxSxG type [11,16] with the central serine being part of the catalytic triad. This lipase motif is indicative of a ⁄ b hydrolase family members [12]. Hydrophobicity predictions [17] indicate a pronounced hydrophobic region in the central domain, consisting of amino acids 154–177 with the core region 164LLILIEPVVI173 (Fig. 1C). By homology searches with other prokaryotic and eukaryotic hydrolases (not shown) using profile hidden Markov models [18], we identified a conserved histidine that is probably part of the catalytic triad of the active site (Fig. 1B). The third member of the catalytic triad could not be identified by sequence-based searches. PTS1-dependent targeting of Lpx1p to peroxisomes The majority of the Lpx1p in a cell homogenate was pelleted at 25 000 g, consistent with an organellar localization of the protein (Fig. 2A). In this experiment, more of the peroxisomal soluble thiolase Fox3p (EC 2.3.1.x) than of Lpx1p appears to be present in the supernatant. This is probably due to partial peroxisome rupture during preparation, and might indicate that Lpx1p, in contrast to Fox3p, is loosely associated with the peroxisomal membrane. The peroxisomal localization of Lpx1p had been demonstrated indirectly by immuno-colabelling of a heterozygous C-terminally Protein A-tagged version of Lpx1p in a diploid strain [10]. Peroxisomal localization under these conditions would depend on the presence of copies of Lpx1p that are not blocked by a C-terminal tag, and by the interaction of Lpx1p with itself (piggyback import). We wished to analyse whether Lpx1p directly localized to peroxisomes, and cloned LPX1 for expression from a yeast shuttle plasmid using an N-terminal GFP tag. This fusion protein was localized to peroxisomes in a Dlpx1 deletion strain (Fig. 2B), indicating that Lpx1p by itself targets to peroxisomes. Peroxisomal localization of Lpx1p was abolished when Lpx1p was expressed with a C-terminal tag (Fig. 2C), indicating that the C-terminus has to be free for Pex5p-dependent import. Peroxisomal localization was abolished in the absence of Pex5p (Fig. 2C), and was not affected by the absence of Pex7p (Fig. 2C), indicating that its targeting to peroxisomes is dependent on the PTS1 pathway. We confirmed the peroxisomal localization of Lpx1p by subcellular fractionation. On a sucrose density gradient, GFP–Lpx1p co-migrated with Fox3p (alternative FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS 505 Peroxisomal lipase Lpx1p S. Thoms et al. Fig. 1. Identification of Lpx1p from Saccharomyces cerevisiae peroxisomes by proteomics. (A) Isolation of putative peroxisomal proteins by preparative chromatographic separation. Low saltextractable peroxisomal proteins were solubilized by SDS and separated by reverse phase HPLC. Polypeptides of selected fractions were separated by SDS–PAGE and visualized by Coomassie blue staining. Only the first 13 lanes of the HPLC profile are shown [15]. The band marked by an asterisk contains the peroxisomal proteins Lpx1p (predicted molecular mass 44 kDa) and Aat2p (predicted molecular mass 44 kDa) in HPLC fraction 7 at a molecular mass of approximately 45 kDa. (B) Alignment of the LPX1 gene with a Mycoplasma genitale (Mg) gene encoding a putative esterase ⁄ lipase (AAC71532) and with the putative triacylglycerol lipase AAB96044 from Mycoplasma pneumoniae (Mp). Identical amino acids are indicated by an asterisk and similar amino acids are indicated by a colon and full point, depending on degree of similarity. The conserved GxSxG lipase motif is shaded in grey. The lipase motif contains the putative active-site serine. The arrowhead indicates the probable active-site histidine, as determined from alignments using eukaryotic esterase lipase family members (not shown). The third member of the catalytic triad could not be identified by sequence-based analysis. (C) Hydropathy plot of Lpx1p. A Kyte–Doolittle plot was calculated with window size of 11. Values > 1.8 may be regarded as highly hydrophobic regions. (D) Termini of all four QKL proteins from S. cerevisiae. Only Lpx1p is predicted to target to peroxisomes. Positions relative to the (putative) PTS1 are indicated. Grey boxes, lysine in position -1 and valine in position -5 are probably required to target Lpx1p to peroxisomes. Lpx1p was identified from low-salt-extractable membranes (Fig. 1A), and the amount of Lpx1p that is not membrane-associated or found in the non-peroxisomal low-density fractions (Fig. 2D; fractions 19–29) is low compared to Fox3p. Although the QKL C-terminus of Lpx1p does not match the PTS1 consensus [SAC][KRH][LM], a QKL terminus is able to target a test substrate to peroxisomes [19]. Lpx1p is one of four S. cerevisiae proteins that end in QKL (Fig. 1D), and is probably the only one that is localized to peroxisomes. Self-interaction of Lpx1p name: Pot1p), with Pex11p, and with the catalase (EC 1.11.1.6) activity peak in the same density fraction at about 1.225 gÆcm)3 (fraction 10) (Fig. 2D). The activity of the mitochondrial marker fumarase (EC 4.2.1.2) together with the mitochondrial Mir1p showed a clearly separate peak at a density of 1.192 gÆcm)3 in fraction 14 (Fig. 2D). 506 C-terminally tagged Lpx1p localizes only to peroxisomes when endogenous copies of the protein are present [10]. This suggests piggyback import of Lpx1p, which, in turn, would rely on self-interaction of Lpx1p. We tested this hypothesis by two-hybrid analysis of LPX1. Neither the fusion of Lpx1p with the GAL4 binding domain nor its fusion with the activation domain were auto-activating (Fig. 3A). The strains expressing both fusions exhibit a strong two-hybrid interaction signal, exceeding that of the control PEX11 with PEX19 (Fig. 3A). Because complex formation FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS S. Thoms et al. Peroxisomal lipase Lpx1p Fig. 2. Localization and PTS1-dependent targeting of Lpx1p to peroxisomes. (A) Immunological detection of GFP–Lpx1p in a sedimentation experiment. A cell-free homogenate (T) was separated into supernatant (S) and an organelle-containing pellet fraction (P) by centrifugation at 25 000 g (30 min). Amounts corresponding to equal T content of each fraction were analysed by SDS–PAGE and western blotting with antibodies against GFP and the peroxisomal marker protein oxoacyl CoA thiolase, Fox3p (alternative name: Pot1p). (B) Lpx1p is localized to peroxisomes. Coexpression of PTS2-dsRed and GFP–Lpx1p in yeast cells. Cells were grown on ethanol to induce the expression of PTS2dsRed. (C) Import of Lpx1p into peroxisomes is dependent on Pex5p and independent of Pex7p. Lpx1p was expressed as either a C-terminal fusion (top images) or N-terminal fusion (bottom images) with GFP. In the Dpex5 deletion mutant, Lpx1p cannot be imported into peroxisomes, irrespective of the position of the tag (right). Deletion of PEX7 does not influence Lpx1p targeting if the PTS1 is not blocked by GFP (top left). GFP fusion proteins that are not targeted to peroxisomes mislocalize to the cytosol. Bar = 2 lm. (D) Sucrose density gradient analysis of GFP–LPX1-transformed yeast. A cell-free organelle sediment from oleate induced cells was analysed on a density gradient with sucrose concentrations form 32 to 54% w ⁄ v. Individual fractions were analysed for catalase activity (peroxisomal marker) and fumarase activity (mitochondrial marker). In addition, the presence of GFP–Lpx1p, Fox3p, Pex11p (peroxisomal membrane protein) and Mir1p (mitochondrial phosphate carrier) was tested by western blotting and immunodetection. might play a significant role in peroxisomal (piggyback) protein import [20], we determined the size of the Lpx1p complex by gel filtration of cell lysates of oleate-induced cells on a Superdex 200 column. We found that the majority of Lpx1p is not present in high-molecularmass complexes, but elutes at molecular masses corresponding to monomers, dimers and trimers (Fig 3B). The two-hybrid interaction probably reflects the complex formation. However, our identification of lowmolecular-mass complexes of Lpx1 does not exclude the possibility that higher-molecular-mass complexes are transiently formed during topogenesis of the protein. on oleate as the only carbon source (Fig. 4A). To determine the influence of Lpx1p on peroxisome biogenesis in more detail, post-nuclear supernatants were prepared from wild-type and Dlpx1 strains. The postnuclear supernatants were analysed by Optiprep gradient analysis and subsequent tests of gradient fractions for peroxisomal catalase and mitochondrial cytochrome c oxidase (EC 1.9.3.l; Fig. 4B). None of these marker proteins indicated a significant change in the abundance or density of peroxisomes or mitochondria, suggesting that peroxisomal and mitochondrial biogenesis remain functional after deletion of the LPX1 gene. Lpx1p is not required for peroxisome biogenesis Lipase activity of Lpx1p Having shown that Lpx1p is targeted to peroxisomes by the soluble PTS1 receptor, we wished to determine whether Lpx1p is required for the biogenesis of peroxisomes. We first tested the Dlpx1 knockout for growth on oleate. However, Lpx1p is dispensable for growth Characteristic GxSxG motifs and similarities with a ⁄ b hydrolases in the predicted protein sequence suggest that Lpx1p is an esterase, possibly a lipase [11,12,16]. To directly investigate Lpx1p, we expressed the full-length protein as a fusion protein with a C-ter- FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS 507 Peroxisomal lipase Lpx1p S. Thoms et al. Fig. 3. Lpx1p interacts with itself. (A) Two-hybrid assay. Full-length Lpx1p was fused to the GAL4 binding or activation domain and coexpressed in a yeast strain with Escherichia coli b-galactosidase under the control of a GAL4-inducible promotor. b-galactosidase activity was measured in lysates of doubly transformed strains. No signal was obtained when LPX1 was combined with empty vectors. Positive control: interaction of Pex19p with Pex11p. (B) Size-exclusion chromatography of a wild-type cell lysate of oleate-induced cells. The lysate was fractionated by gel filtration on a Superdex 200 column and tested by immunoblotting with anti-Lpx1p antiserum. The molecular masses indicated were interpolated from a calibration curve and correspond well with monomeric, dimeric and trimeric forms of Lpx1p. The relative distribution of the three forms was quantified using NIH Image (National Institutes of Health, Bethesda, MD, USA). The elution volume is indicated in millilitre. minal hexahistidine tag in Escherichia coli and purified the protein using immobilized metal-ion affinity chromatography (Fig. 5A). The protein was further purified by gel filtration on a Superdex 200 column (Fig. 5B). Gel filtration indicated the propensity of Lpx1p to oligomerize in vitro, albeit to a much lower extent than in yeast whole-cell lysates (compare Figs 3B and 5B). Purified protein was used for the generation of polyclonal antibodies in rabbit. Antisera recognized a protein of about 43 kDa, indicating that the antiserum is specific for Lpx1p. We used these antibodies to confirm that the endogenous yeast Lpx1p is induced by oleic acid (Fig. 5A). To analyse the enzyme activity of Lpx1p, we assayed the E. coli-expressed protein for esterase activity, using p-nitrophenyl butyrate (PNB) as the test substrate. PNB can be hydrolysed by esterases, yielding free p-nitrophenol, which can be determined photometrically at 410 nm. Lpx1p hydrolysed the test substrate with a KM of 6.3 lm and Vmax of 0.17 lmolÆs)1 (Table 1). Lpx1p is strongly induced by oleic acid, regulated by stress-associated transcription factors [21], and aligns 508 Fig. 4. Absence of pex phenotype in a Dlpx1 deletion. (A) Growth on plates with oleate as the only carbon source. Wild-type, Dlpx1 or Dpex1 control stains were spotted in equal cell numbers in series of 10-fold dilutions on oleate or ethanol plates. Absence of growth and oleic acid consumption (halo formation) indicates a peroxisomal defect. Control: growth assay on ethanol. (B) Optiprep density gradient centrifugation analysis of postnuclear supernatants prepared from oleate-induced wild-type and Dlpx1 strains. All fractions were analysed using catalase (peroxisome) and cytochrome c oxidase (mitochondria) enzyme assays. The peroxisomal and mitochondrial densities were not measurably altered by LPX1 deletion. with human epoxide hydrolases (EC 1.14.99.x; not shown). We found that Lpx1 hydrolysed the epoxide hydrolase substrate [22] 4-nitrophenyl-trans-2,3-epoxy3-phenylpropyl carbonate (NEPC) (data not shown), but we consider that this activity is non-specific, because it could not be blocked by the specific epoxide hydrolase inhibitor N,N’-dicyclohexylurea (DCU) (data not shown). To test for lipase activity, we used 1,2-dioleoyl-3(pyren-1-yl)decanoyl-rac-glycerol (DPG) as a substrate. DPG contains the eximer-forming pyrene decanoic acid as one of the acyl residues. Upon cleavage, the free pyrene decanoic acid shows reduced eximer fluorescence. Lpx1p exerts lipase activity towards DPG of 5.6 pmolÆh)1Ælg)1 (Table 1). For comparison, we measured the lipase activity of commercial yeast Candida rugosa lipase towards DPG and found an FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS S. Thoms et al. Peroxisomal lipase Lpx1p Fig. 5. Protein expression, antibody generation and oleate induction of Lpx1p. Expression of Lpx1p in Escherichia coli. (A) Lpx1p was expressed as a fusion protein with a C-terminal hexahistidine tag and purified by His-trap chromatography. The purified Lpx1p (lane 1) was used to generate polyclonal antibodies in rabbit that recognize the purified recombinant protein (lane 4). Endogenous Lpx1p in whole yeast lysates is recognized only after induction of peroxisomes and Lpx1p by oleate (lane 2 versus lane 3). Molecular masses are shown in kDa. (B) Second purification step: gel filtration on Superdex 200 column. The elution profile indicates that most of the protein behaves as a monomer, but a small proportion forms dimers and trimers. Table 1. Esterase, lipase, and phospholipase activity of Lpx1p. Esterase activity was measured using PNB (p-nitrobutyrate) as a substrate. KM and Vmax values were calculated using Michaelis– Menten approximations. Lipase activity was determined using DPG as a substrate. Activity was measured from two independent protein preparations in triplicate. Candida rugosa lipase (CRL) was used as a positive control for lipase measurement. (Pancreas) lipase activity assays used DGR in a coupled enzyme assay as a substrate. Phospholipase C and D (PLC and PLD) activities were measured in coupled enzyme assays using phosphatidylcholine (PC). Phospholipase A measurements used BPC (bis-BODIPY-FL C11-PC) as a test substrate. Porcine pancreas lipase (PPL) was used as a control. Enzyme Substrate Activity Lpx1p PNB Acyl esterase Lpx1p DPG CRL DPG Lpx1p Lpx1p Lpx1p Lpx1p PPL DGR PC PC BPC BPC (Triacylglycerol) lipase (Triacylglycerol) lipase (Pancreas) lipase PLC PLD PLA PLA Activity parameters (pmolÆh)1Ælg)1) KM 6.3 lM; Vmax 0.17 lmolÆs)1 5.6 ± 1.5 2.0 ± 0.1 Below detection limit Below detection limit Below detection limit 7.9 195 activity of 2.0 pmolÆh)1Ælg)1 under the same assay conditions (Table 1). We sought to confirm lipase activity by testing Lpx1p in a clinical assay for pancreatic lipase. The assay uses the substrate 1,2-O-dilauryl-rac-glycero3-glutaric acid (6-methyl resorufin) ester (DGR) in a desoxycholate-containing buffer. Lpx1p did not hydrolyse this substrate under the assay conditions (Table 1). Next we tested for phospholipase C activity in a coupled enzyme assay with phosphatidylcholine as the substrate. In this assay, phospholipase C converts phosphatidylcholine to phosphocholine and diacylglycerol. Alkaline phosphatase hydrolyses phosphocholine to form choline, which is then oxidized by choline oxidase to betaine and hydrogen peroxide. The latter, in the presence of horseradish peroxidase, reacts with 10-acetyl-3,7-dihydrophenoxazine to form fluorescent resorufin. This assay, as well as a similar assay for phospholipase D, gave negative results for Lpx1p (Table 1). Finally, we tested phospholipase A (EC 3.1.1.4) activity using the substrate 1,2-bis-(4,4-difluoro-5,7dimethyl-4-bora-3a,4a-diaza-sindacene-3-undecanoyl)sn-glycero-3-phosphocholine (bis-BODIPY-FL C11-PC, BPC). BPC is a glycerophosphocholine with BODIPY dye-labeled sn-1 and sn-2 C11 acyl chains. Cleavage reduces dye quenching and leads to a fluorecence increase at 530 nm upon excitation at 488 nm. Lpx1p exerts phospholipase A activity of 7.9 pmolÆh)1Ælg)1. As a control enzyme, we used commercial porcine pancreas lipase, which hydrolysed 195 pmolÆh)1Ælg)1. In summary, Lpx1p shows acyl esterase, lipase and phospholipase A activity towards PNB, DPG and BPC, respectively. Altered peroxisome morphology in deletion mutants of LPX1 Lastly, we analysed electron microscopic (EM) images of knockouts of LPX1. To our surprise, a large number of Dlpx1 peroxisomes showed an abnormal morphology. The peroxisomes appear vesiculated FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS 509 Peroxisomal lipase Lpx1p S. Thoms et al. (Fig. 6B), and either contain intraperoxisomal vesicles or their membrane is grossly invaginated. On average, one vesiculated peroxisome is visible in every fifth mutant cell (Fig. 6E). When the average number of altered peroxisomes is counted, we find that every third peroxisome shows this vesiculation phenotype (Fig. 6D). This is a high percentage, considering the fact that the peroxisomes were viewed in thin microtome sections. In three dimensions, every single peroxisome might contain a vesicular membrane or indentation that escapes notice in two-thirds of the ‘two-dimensional’ sections. The average number of peroxisomes per cell is insignificantly increased in Dlpx1 (2.95 versus 2.76, Fig. 6C). Wild-type cells did not contain any vesiculated peroxisome (Fig. 6A,D,E). The drastic phenotype of Dlpx1 is reminiscent of the peroxisomal morphology found in peroxisomal disorders. Discussion Lpx1p is a peroxisomal protein with an unusual PTS1 Fig. 6. Peroxisome morphology phenotype of the Dlpx1 deletion. Absence of LPX1 leads to drastic peroxisomal vesiculation or invagination. Electron microscopic images of cells from (A) wild-type and (B) Dlpx1. All cells were grown on medium with 0.1% oleic acid. Peroxisomes are marked by arrowheads. Bar = 2 lm. (C) Comparison of per cell peroxisome numbers in wild-type and Dlpx1 strains. (D) Average number of vesicles per peroxisome (wild-type, n = 94; Dlpx1, n = 142). In Dlpx1, about every third peroxisome contains a vesicle. (E) Percentage of cells with vesicle-containing peroxisomes. Roughly one in five Dlpx1 cells carries peroxisomes with a vesicle or invaginations (wild-type, n = 34; Dlpx1, n = 48). px, peroxisome(s). 510 LPX1 is one of the most strongly induced genes following a shift from glucose to oleate, as determined by serial analysis of gene expression (SAGE) experiments [9]. The oleate-induced increase in mRNA abundance is abolished in the Dpip2 Doaf1 double deletion strain, indicating that its induction is dependent on the transcription factor pair Pip2p and Oaf1p [9]. The Lpx1p protein itself is induced by oleic acid as determined using a Protein A tag [10] or by use of an antibody raised against Lpx1p (see Results). Lpx1p does not conform to the general PTS1 consensus. The other three QKL proteins in S. cerevisiae are probably not peroxisomal (Fig. 1D): Efb1p (systematic name: Yal003wp) is the elongation factor EF-1b [23], Rpt4p (Yor259cp) is a mostly nuclear 19S proteasome cap AAA protein [24], and Tea1p (Yor337wp) is a nuclear Ty1 enhancer activator [25]. However, QKL is sufficient to sponsor Pex5p binding [19]. Why are these QKL proteins not imported into peroxisomes? This is probably due to the upstream sequences. Lpx1p has a lysine at position -1 (relative to the PTS1 tripeptide) and a hydrophobic amino acid at position -5 (Fig. 1D). These features promote Pex5p binding and are not found in the other three QKL proteins (Fig. 1D) [19]. Our views were confirmed by applying a PTS1 prediction algorithm (http:// mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp) [26], which predicted peroxisomal localization for Lpx1p only of the four proteins listed in Fig. 1D. FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS S. Thoms et al. Lipase activity and cellular function of Lpx1p Lpx1p could be involved in various processes: (a) detoxification and stress response, (b) lipid mobilization, or (c) peroxisome biogenesis. As Lpx1p expression may be regulated by Yrm1p and Yrr1p [21], a transcription factor pair that mediates pleiotropic drug resistance effects, we speculate that Lpx1p is required for a multidrug resistance response that did not show a phenotype in our experiments. We could, however, exclude epoxide hydrolase activity for Lpx1p, because hydrolysis of the epoxide hydrolase test substrate was not affected by a specific epoxide hydrolase inhibitor. We investigated the dimerization of Lpx1p in the context of piggyback protein import into peroxisomes. Self-interaction (dimerization) is frequently found in regulation of the enzymatic activity of other lipases such as C. rugosa lipase or human lipoprotein lipase [27,28]. The putative active-site serine of Lpx1p is located next to the region of highest hydropathy, suggesting that Lpx1p is a membrane-active lipase that contributes to metabolism or the membrane shaping of peroxisomes. Peroxisomes are sites of lipid metabolism. It is thus not surprising to find a lipase associated with peroxisomes. Our experiments show that Lpx1p has triacylglycerol lipase activity; however, activities towards the artificial test substrates DPG and DGR were low. Our evidence for phospholipase A activity of the enzyme, together with the EM phenotype, suggest that Lpx1 has a more specialized role in modifying membrane phospholipids. Recently, a mammalian group VIB calcium-independent phospholipase A2 (iPLA2c) was identified that possesses a PTS1 SKL and a mitochondrial targeting signal [29,30]. The enzyme is localized in peroxisomes and mitochondria, and is involved, among others, in arachinoic acid and cardiolipin metabolism [31,32]. Knockout mice of iPLA2c show mitochondrial ⁄ cardiological phenotypes [33]. It will be exciting to determine whether human iPLA2c and yeast Lpx1p are functionally related. We have provided evidence that peroxisomes are still functional in the absence of LPX1. This suggests a non-essential metabolic role for Lpx1p in peroxisome function. The morphological defect found in electron microscopic images of a deletion of Lpx1p (peroxisomes containing inclusions or invaginations) is symptomatic of a yeast peroxisomal mutant, and is reminiscent of the phenotypes found in human peroxisomal disorders [4,5]. Out data suggest that Lpx1p is required to determine the shape of peroxisomes. Peroxisomal lipase Lpx1p Experimental procedures Strains and expression cloning The S. cerevisiae strains BY4742, BY4742 Dyor084w, BY4742 Dpex5, BY4742 Dpex7 and BY4742 Dpex1 were obtained from EUROSCARF (Frankfurt, Germany). S. cerevisiae strain BJ1991 (Mata leu2 trp1 ura3-251 prb1-1122 pep4-3) has been described previously [34]. Genomic S. cerevisiae DNA was used as a PCR template for PCR. For construction of pUG35-LPX1 (LPX1–GFP), PCR-amplified YOR084w (primers 5¢-GCTCTAGAATG GAACAGAACAGGTTCAAG-3¢ and 5¢-CGGAATTCCA GTTTTTGTTTAGTCGTTTTAAC-3¢) was subcloned into EcoRV-digested pBluescript SK+ (Stratagene, La Jolla, CA, USA), and then introduced into the XbaI and EcoRI sites of pUG35 (HJ Hegemann, Düsseldorf, Germany). For construction of pUG36-LPX1 (GFP–LPX1), PCR-amplified YOR084w (primers 5¢-GAGGATCCATGGAACAGAACA GGTTCAAG-3¢ and 5¢-CGGAATTCTTACAGTTTTTGT TTAGTCGTTTTAAC-3¢) was subcloned into EcoRVdigested pBluescript SK+, and then cloned into the BamHI and EcoRI sites of pUG36 (HJ Hegemann). pET21d-LPX1 was constructed by introducing PCRamplified YOR084w (primers 5¢-GAATCCATGGAACAG AACAGGTTCAA-3¢ and 5¢-CGGTACCGCGGCCGCCA GTTTTTGTTTAGTCGTTTT-3¢) into the NcoI and NotI sites of pET21d (EMD Chemicals, Darmstadt, Germany). For construction of pPC86-LPX1 and pPC97-LPX1, YOR084w was amplified using primers 5¢-CCCGGGAAT GGAACAGAACAGGTTCAAG-3¢ and 5¢-AGATCTTTA CAGTTTTTGTTTAGTCGTTTT-3¢, and introduced into pGEM-T (Promega, Mannheim, Germany). The ORF was excised using XmaI and BglII, and introduced into pPC86 and pPC97 [35]. All constructs were confirmed by DNA sequencing. pPTS2-DsRed has been described previously [36]. Image acquisition Samples were fixed with 0.5% w ⁄ v agarose on microscopic slides. Fluorescence microscopic images were recorded on an Axioplan2 microscope (Zeiss, Köln, Germany) equipped with an aPlan-FLUAR 100 x ⁄ 1.45 oil objective and an AxioCam MRm camera (Zeiss) at room temperature. If necessary, contrast was linearly adjusted using the image acquisition software Axiovision 4.2 (Zeiss). Protein purification and antibody generation Lpx1p was expressed from pET21d-LPX1 in BL21(DE3) E. coli. Cells were harvested by centrifugation (SLA3000, 4000 g, 15 mins), and resuspended in buffer P (1.7 mm potassium dihydrogen phosphate, 5.2 mm disodium hydrogen phosphate, pH 7.5, 150 mm sodium chloride) containing FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS 511 Peroxisomal lipase Lpx1p S. Thoms et al. a protease inhibitor mix (8 lm antipain-dihydrochloride, 0.3 lm aprotinin, 1 lm bestatin, 10 lm chymostatin, 5 lm leupeptin, 1.5 lm pepstatin, 1 mm benzamidin, and 1 mm phenylmethane sulfonylfluoride) and 50 lgÆmL)1 lysozyme, 22.5 lgÆmL)1 DNAse I and 40 mm imidazole. Cells were sonicated 20 times for 20 s each using a 250D Branson digital sonifier (Danbury, CT, USA) with an amplitude setting of 25%. After removal of cell debris (SS34, 27 000 g, 45 min) the supernatant was clarified by 0.22 lm filtration (Sarstedt, Nümbrecht, Germany) and loaded on Ni-Sepharose columns (GE Healthcare, Munich, Germany) equilibrated with buffer W (buffer P containing 300 mm sodium chloride, 1 mm dithiothreitol, 40 mm imidazole). The column was washed in buffer W until no further protein was eluted. Recombinant Lpx1p was eluted by a continuous 40–500 mm imidazole gradient based on buffer W. Peak fractions (identified by SDS–PAGE) were pooled and concentrated using VivaSpin concentrators (30 kDa cutoff, Sartorius, Göttingen, Germany). Lpx1p was further purified by gel-filtration chromatography. Protein was stored at 0 C. For the production of polyclonal antibodies, gel bands corresponding to 150 lg protein were excised and used for rabbit immunization (Eurogentec, Seraing, Belgium). of 200 lL at 37 C. Hydrolysis of DPG was followed in 96-well plates at 460 nm with 360 nm excitation using a Sirius HT fluorescence plate reader (MWG Biotech, Ebersberg, Germany). Lipase activity towards DPG was measured in assay setups containing 2–10 lg Lpx1p (from two independent protein preparations), with C. rugosa triacylglycerol lipase (Lipase AT30 Amano, 1440 UÆmg)1, Sigma-Aldrich) as a control. Phospholipase A activity was measured using bisBODIPY FL C11-PC (Molecular Probes ⁄ Invitrogen, Eugene, OR, USA) as the substrate. The assay setup contained 70 lg Lpx1p in 50 lL assay buffer (50 mm Tris, 100 mm sodium chloride, 1 mm calcium carbonate, pH 8.9) together with 50 lL substrate-loaded liposomes. Liposomes were prepared by injecting 90 lL of an ethanolic mixture of 3.3 mm dioleyl phosphatidylcholine (Avanti Polar Lipids, Alabaster, AL, USA), 3.3 mm dioleyl phosphatidylglycerol (Avanti Polar Lipids) and 0.33 mm bis-BODIPY FL C11PC into 5 ml assay buffer. Substrate turnover was measured at 528 nm emission after 488 nm excitation. Activity was calculated from the initial velocity. Porcine pancreas phospholipase A2 (Fluka ⁄ Sigma-Aldrich, Buchs, Swizerland) was used as a control. Density gradient centrifugation Size-exclusion chromatography For analysis of endogenous Lpx1p by gel filtration, 5 mL of a glass bead lysate of oleate-induced BY4742 wild-type cells in buffer A (buffer P, pH 7.3, 300 mm sodium chloride) with a protease inhibitor mix were injected into a HiLoad 16 ⁄ 60 Superdex 200 prepgrade column (GE Healthcare) and eluted using buffer A at a flow rate of 1 mL)1Æmin and a fraction size of 2 mL. Fractions were analysed by SDS–PAGE and Western blotting. A 500 lL aliquot of the concentrated Ni-Sepharose eluate of Lpx1p from E. coli expression was purified in the same buffer under the same conditions. For estimation of Lpx1p complex sizes, molecular masses were interpolated from a calibration curve generated using ovalbumin (45 kDa), carboanhydrase (29 kDa), trypsin inhibitor (20.1 kDa), lactalbumin (14.2 kDa) and aprotinin (6.5 kDa) as molecular mass standards. Enzyme assays Esterase activity was determined using 0.5 mm p-nitrophenyl butyrate (Sigma-Aldrich, Seelze, Germany) in NaCl ⁄ Pi (pH 7.4) in a total volume of 200 lL at 37 C. The amount of free p-nitrophenol was determined at 410 nm in 96-well plates. Michaelis–Menten kinetics were analysed using GraphPad Prism4 (Graph Pad Software, San Diego, CA, USA). Lipase activity was determined using 0.5 mm DPG (Marker Gene Technologies, Eugene, OR, USA) in 0.1 m glycine, 19 mm sodium deoxicholate, pH 9.5, in a total volume 512 Gradient centrifugation was carried out essentially as described previously [37]. Briefly, oleate-induced yeast cells were converted to spheroblasts using 25 UÆg)1 Zymolyase 100T (MP Biomedicals, Illkirch, France). Spheroblasts were gently ruptured by Potter–Elvehjem homogenization, and centrifuged at low speed (3 · 10 min at 600 g) to generate postnuclear supernatants. These supernatants, containing 5 mg protein, were loaded on a 32–54% sucrose gradient (Fig. 2D) or an Optiprep gradient (Fig. 4B) and centrifuged for 3 h at 19 000 g (Sorvall SV288, 19 000 rpm, 4 C). The gradient was fractionated into about 29 fractions of 1.2 mL. Fractions were analysed using enzyme assays for oxoacyl CoA thiolase, catalase, fumarase and cytochrome c oxidase [37]. Other methods Mass spectrometry and high-pressure lipid chromatography have been described previously [14,15,38,39]. Subcellular fractionation, yeast two-hybrid assays and electron microscopy were carried out as described previously [37]. Acknowledgements We thank Elisabeth Becker, Monika Bürger and Uta Ricken for technical assistance. We thank Sabine Weller and Hartmut Niemann for reading the manuscript. We extend our thanks to three anonymous reviewers FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS S. Thoms et al. who helped to improve the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (Er178 ⁄ 2-4) and by the Fonds der Chemischen Industrie. References 1 Wanders RJ & Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75, 295–332. 2 Weller S, Gould SJ & Valle D (2003) Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet 4, 165–211. 3 Thoms S & Erdmann R (2005) Import of proteins into peroxisomes. In Protein Movement Across Membranes (Eichler J, ed.), pp. 125–134. Landes Bioscience ⁄ Springer, New York, NY. 4 Motley A, Hettema E, Distel B & Tabak H (1994) Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol 125, 755–767. 5 Funato M, Shimozawa N, Nagase T, Takemoto Y, Suzuki Y, Imamura Y, Matsumoto T, Tsukamoto T, Kojidani T, Osumi T et al. (2006) Aberrant peroxisome morphology in peroxisomal b-oxidation enzyme deficiencies. Brain Dev 28, 287–292. 6 Thoms S & Erdmann R (2006) Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta 1763, 1620–1628. 7 Platta HW & Erdmann R (2007) The peroxisomal protein import machinery. FEBS Lett 581, 2811–2819. 8 Petriv OI, Tang L, Titorenko VI & Rachubinski RA (2004) A new definition for the consensus sequence of the peroxisome targeting signal type 2. J Mol Biol 341, 119–134. 9 Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B et al. (1999) Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell 10, 1859– 1872. 10 Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA & Aitchison JD (2002) Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol 158, 259–271. 11 Voss H, Benes V, Andrade MA, Valencia A, Rechmann S, Teodoru C, Schwager C, Paces V, Sander C & Ansorge W (1997) DNA sequencing and analysis of 130 kb from yeast chromosome XV. Yeast 13, 655–672. 12 Schrag JD & Cygler M (1997) Lipases and a ⁄ b hydrolase fold. Methods Enzymol 284, 85–107. 13 Schafer H, Nau K, Sickmann A, Erdmann R & Meyer HE (2001) Identification of peroxisomal membrane Peroxisomal lipase Lpx1p 14 15 16 17 18 19 20 21 22 23 24 25 26 27 proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22, 2955–2968. Erdmann R & Blobel G (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol 128, 509–523. Geisbrecht BV, Schulz K, Nau K, Geraghty MT, Schulz H, Erdmann R & Gould SJ (1999) Preliminary characterization of Yor180Cp: identification of a novel peroxisomal protein of Saccharomyces cerevisiae involved in fatty acid metabolism. Biochem Biophys Res Commun 260, 28–34. Brenner S (1988) The molecular evolution of genes and proteins: a tale of two serines. Nature 334, 528–530. Kyte J & Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157, 105–132. Soding J (2005) Protein homology detection by HMM– HMM comparison. Bioinformatics 21, 951–960. Lametschwandtner G, Brocard C, Fransen M, Van Veldhoven P, Berger J & Hartig A (1998) The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem 273, 33635–33643. Gould SJ & Collins CS (2002) Opinion: peroxisomalprotein import: is it really that complex? Nat Rev Mol Cell Biol 3, 382–389. Lucau-Danila A, Delaveau T, Lelandais G, Devaux F & Jacq C (2003) Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J Biol Chem 278, 52641–52650. Mateo C, Archelas A & Furstoss R (2003) A spectrophotometric assay for measuring and detecting an epoxide hydrolase activity. Anal Biochem 314, 135–141. Hiraga K, Suzuki K, Tsuchiya E & Miyakawa T (1993) Cloning and characterization of the elongation factor EF-1b homologue of Saccharomyces cerevisiae. EF-1b is essential for growth. FEBS Lett 316, 165–169. McDonald HB & Byers B (1997) A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol 137, 539–553. Gray WM & Fassler JS (1996) Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol Cell Biol 16, 347– 358. Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A & Eisenhaber F (2003) Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J Mol Biol 328, 581–592. Pernas MA, Lopez C, Pastrana L & Rua ML (2001) Purification and characterization of Lip2 and Lip3 FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS 513 Peroxisomal lipase Lpx1p 28 29 30 31 32 33 514 S. Thoms et al. isoenzymes from a Candida rugosa pilot-plant scale fedbatch fermentation. J Biotechnol 84, 163–174. Goldberg IJ & Merkel M (2001) Lipoprotein lipase: physiology, biochemistry, and molecular biology. Front Biosci 6, D388–D405. Yang J, Han X & Gross RW (2003) Identification of hepatic peroxisomal phospholipase A(2) and characterization of arachidonic acid-containing choline glycerophospholipids in hepatic peroxisomes. FEBS Lett 546, 247–250. Mancuso DJ, Jenkins CM, Sims HF, Cohen JM, Yang J & Gross RW (2004) Complex transcriptional and translational regulation of iPLAc resulting in multiple gene products containing dual competing sites for mitochondrial or peroxisomal localization. Eur J Biochem 271, 4709–4724. Murakami M, Masuda S, Ueda-Semmyo K, Yoda E, Kuwata H, Takanezawa Y, Aoki J, Arai H, Sumimoto H, Ishikawa Y et al. (2005) Group VIB Ca2+-independent phospholipase A2c promotes cellular membrane hydrolysis and prostaglandin production in a manner distinct from other intracellular phospholipases A2. J Biol Chem 280, 14028–14041. Mancuso DJ, Han X, Jenkins CM, Lehman JJ, Sambandam N, Sims HF, Yang J, Yan W, Yang K, Green K et al. (2007) Dramatic accumulation of triglycerides and precipitation of cardiac hemodynamic dysfunction during brief caloric restriction in transgenic myocardium expressing human calcium-independent phospholipase A2gamma. J Biol Chem 282, 9216–9227. Mancuso DJ, Sims HF, Han X, Jenkins CM, Guan SP, Yang K, Moon SH, Pietka T, Abumrad NA, Schlesinger PH et al. (2007) Genetic ablation of calcium-independent phospholipase A2c leads to alterations in 34 35 36 37 38 39 mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J Biol Chem 232, 34611–34622. Woolford CA, Noble JA, Garman JD, Tam MF, Innis MA & Jones EW (1993) Phenotypic analysis of proteinase A mutants. Implications for autoactivation and the maturation pathway of the vacuolar hydrolases of Saccharomyces cerevisiae. J Biol Chem 268, 8990–8998. Chevray PM & Nathans D (1992) Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA 89, 5789–5793. Stein K, Schell-Steven A, Erdmann R & Rottensteiner H (2002) Interactions of Pex7p and Pex18p ⁄ Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol Cell Biol 22, 6056–6069. Schafer A, Kerssen D, Veenhuis M, Kunau WH & Schliebs W (2004) Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol Cell Biol 24, 8895–8906. Schäfer H, Nau K, Sickmann A, Erdmann R & Meyer HE (2001) Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22, 2955–2968. Immler D, Gremm D, Kirsch D, Spengler B, Presek P & Meyer HE (1998) Identification of phosphorylated proteins from thrombin-activated human platelets isolated by two-dimensional gel electrophoresis by electrospray ionization–tandem mass spectrometry (ESI-MS ⁄ MS) and liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS). Electrophoresis 19, 1015–1023. FEBS Journal 275 (2008) 504–514 ª 2008 The Authors Journal compilation ª 2008 FEBS Seventh International Meeting on AAA Proteins The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p Harald W. Platta, Mykhaylo O. Debelyy, Fouzi El Magraoui and Ralf Erdmann1 Abteilung für Systembiochemie, Medizinische Fakultät der Ruhr-Universität Bochum, D-44780 Bochum, Germany Biochemical Society Transactions www.biochemsoctrans.org Abstract The discovery of the peroxisomal ATPase Pex1p triggered the beginning of the research on AAA (ATPase associated with various cellular activities) proteins and the genetic dissection of peroxisome biogenesis. Peroxisomes are virtually ubiquitous organelles, which are connected to diverse cellular functions. The highly diverse and adaptive character of peroxisomes is accomplished by modulation of their enzyme content, which is mediated by dynamically operating protein-import machineries. The import of matrix proteins into the peroxisomal lumen has been described as the ATP-consuming step, but the corresponding reaction, as well as the ATPase responsible, had been obscure for nearly 15 years. Recent work using yeast and human fibroblast cells has identified the peroxisomal AAA proteins Pex1p and Pex6p as mechano-enzymes and core components of a complex which dislocates the cycling import receptor Pex5p from the peroxisomal membrane back to the cytosol. This AAA-mediated process is regulated by the ubiquitination status of the receptor. Pex4p [Ubc10p (ubiquitin-conjugating enzyme 10)]-catalysed mono-ubiquitination of Pex5p primes the receptor for recycling, thereby enabling further rounds of matrix protein import, whereas Ubc4p-catalysed polyubiquitination targets Pex5p to proteasomal degradation. Introduction Pex1p (formerly Pas1p), Sec18p [NSF (N-ethylmaleimidesensitive factor)] and Cdc48p (cell division cycle 48 protein) [p97/VCP (valosin-containing protein)] represent the first proteins that were recognized as belonging to a novel family of ATPases [1], the AAA (ATPase associated with various cellular activities) family [2], which later was extended to the family of AAA+ proteins [3]. Belonging to the class of P-loop (phosphate-binding) NTPases, the AAA+ proteins are especially distinguished by the presence of at least one evolutionarily conserved 200–250-amino-acid ATP-binding domain that contains Walker A and B motifs in addition to other structural features, such as the SRH (second region of homology), which distinguishes AAA proteins from other AAA+ proteins [4]. Although the members of the AAA+ family display a high functional diversity, the common function of all seems to be the ability to catalyse reactions that are associated with significant conformational remodelling of substrate proteins or nucleic acids [5]. A lot of detailed information regarding the structure and molecular mechanism of AAA proteins has been accumulated, but our understanding of the molecular function of Key words: ATPase associated with various cellular activities (AAA), peroxin, PEX, protein transport, ubiquitination. Abbreviations used: AAA, ATPase associated with various cellular activities; Cdc48p, cell division cycle 48 protein; NSF, N-ethylmaleimide-sensitive factor; NTD, N-terminal domain; PTS, peroxisomal targeting signal; SRH, second region of homology; Ubc, ubiquitin-conjugating enzyme; VCP, valosin-containing protein. 1 To whom correspondence should be addressed (email [email protected]). Biochem. Soc. Trans. (2008) 36, 99–104; doi:10.1042/BST0360099 Pex1p and Pex6p, the second AAA peroxin in peroxisomal biogenesis [6], remained incomplete for many years. Peroxisomes are single-membrane-bound organelles of virtually all eukaryotic cells, which display a unique variability in their enzyme content and metabolic functions that are adjusted according to the cellular needs. Their matrix harbours at least 50 different enzymes that are linked to diverse biochemical pathways. The β-oxidation of fatty acids and the detoxification of hydrogen peroxide are regarded as the central function of peroxisomes. They are the source of signalling molecules such as jasmonates in plants or lipid-derived ligands for PPARs (peroxisomeproliferator-activated receptors) in humans. Other functions of peroxisomes include the final steps of penicillin biosynthesis in some filamentous fungi, the main reactions of photorespiration in leaf peroxisomes and the synthesis of bile acid and ether lipids such as plasmalogens in mammals, which contribute more than 80 % of the phospholipid content of the white matter in the brain [7]. Because of the central role of peroxisomes in lipid metabolism, they are essential for normal human development and physiology. This is emphasized by a group of genetic disorders collectively referred to as the peroxisome disorders, which, in most cases, lead to death in early infancy [8]. Detailed analysis of the complementation groups finally revealed that the most common cause of peroxisomal biogenesis disorders are mutations in Pex1p [9]. More than 80 % of all patients with Zellweger syndrome, the most severe peroxisome biogenesis disorder, carry mutations in Pex1p or Pex6p [10]. C The C 2008 Biochemical Society Authors Journal compilation 99 100 Biochemical Society Transactions (2008) Volume 36, part 1 Molecular architecture of the peroxisomal AAA complex AAA proteins are characterized by a typical modular architecture as they contain an N-terminal non-ATPase domain which is followed by at least one conserved AAA domain. Each AAA cassette usually contains an ATP-binding site (Walker A) and an ATP-hydrolysis site (Walker B) along with other motifs, such as the SRH [4]. Pex1p and Pex6p are type II AAA proteins, which are characterized by two AAA domains (Figure 1). In both AAA peroxins, the second AAA domain is more conserved than the first one. Interaction and subsequent oligomerization of Pex1p and Pex6p is believed to be initiated in the cytosol and involves their first less conserved AAA domains (D1) [11,12]. Although neither binding nor hydrolysis of ATP at D1 seems to be essential for functionality in both yeast and humans, the interaction of human Pex1p and Pex6p is stimulated by binding of ATP to D1 of human Pex1p and Pex6p [12,13]. Furthermore, ATP binding, but not hydrolysis, at the second AAA cassette (D2) of Pex1p is required for the Pex1p–Pex6p interaction in both systems [11,12]. Pex1p and Pex6p are believed to form heterohexameric structures in the cytosol and at the peroxisomal membrane [12,14–16]. However, it is not clear whether formation of a heteromeric assembly of the AAA peroxins is a prerequisite for their function, as one population of Pex1p does not co-localize with Pex6p in mammalian cells [12,17]. Although the formation of hexameric structures is common to AAA proteins, the formation of heterohexamers has been found in few other cases, such as the m-AAA (matrix AAA) complex, consisting of Yta10p and Yta12p, which is active at the matrix site of the inner mitochondrial membrane, [18] or the six different Rpt ATPases from the 19S proteasome [19]. The recruitment of AAA complexes to peroxisomes is mediated by the tail-anchored peroxisomal membrane proteins Pex15p in Saccharomyces cerevisiae or its functional orthologue Pex26p in human cells via binding of the N-terminal domain of Pex6p, stimulated by ATP binding to the Walker A motif of Pex6p D1 [20,21]. In contrast, the Walker A and B motifs of Pex6p D2 are required for an efficient detachment from Pex15p/Pex26p [12,20,22]. Although Pex15p and Pex26p have been described as adaptor proteins for the N-terminal part of Pex6p, no adaptor has yet been identified for Pex1p. The NTD (N-terminal domain) of murine Pex1p represents the only available crystal structure of the AAA peroxins [23]. The NTD folds into two structurally independent globular subdomains (N- and C-lobe), which comprise an N-terminal double-ψ fold and a C-terminal β-barrel, separated by a shallow groove. Similar grooves were found in the adaptor-binding sites within the NTDs of VCP, NSF and VAT (VCP-like ATPase from Thermoplasma), suggesting functional similarity [23]. The Pex15p-anchored AAA complex itself is part of an even larger protein complex at the peroxisomal membrane, C The C 2008 Biochemical Society Authors Journal compilation the peroxisomal matrix protein import machinery called the importomer (Figure 1) [15]. To conclude, at least in S. cerevisiae, the Pex1p-bound nucleotides seem to influence the Pex1p–Pex6p interaction, while the different nucleotide states of Pex6p regulate the dynamic Pex6p–Pex15p/Pex26p association. The nonconserved domains are responsible for oligomerization, while the conserved domains exhibit the main ATPase activity. Pex1p and Pex6p: peroxins associated with diverse cellular activities? Besides their involvement in peroxisomal biogenesis, the AAA peroxins have been suggested to carry out other functions as well. Human Pex6p has been reported to interact specifically with the nucleocytoplasmatic transcriptional regulators Smad2, Smad3, Smad4 and Smad7 [24]. These proteins are involved in the signalling pathway of the plasma membrane receptor TGFβ (transforming growth factor β), which regulates apoptosis. Furthermore, a suppressor screen for aging defects in mitochondria revealed that Pex6p, but not Pex1p, complements an ATP2-caused import defect into mitochondria, indicating a novel, yet not understood, function of this peroxin in mitochondrial inheritance and senescence [25]. In the context of peroxisomal biogenesis, the different functions discussed are mostly linked to the modulation of membrane dynamics. On the basis of the finding that Pex1p and Pex6p can associate with membranous subcellular structures distinct from mature peroxisomes in the yeasts Pichia pastoris and Yarrowia lipolytica, these peroxins were thought to play a role in lipid or membrane transport [14,26]. Utilizing in vitro fusion experiments, Pex1p and Pex6p were shown to be required for the fusion of five different premature peroxisomal vesicle species in Y. lipolytica [26], a process which might play a role during the maturation of endoplasmic reticulum-derived peroxisomal structures during de novo synthesis of peroxisomes [27]. The still putative functional relevance of the observed phospholipidbinding activity of the murine Pex1p NTD, which has also been described for VCP and NSF, might be linked to this process [28]. Furthermore, the presence of Pex6p and Pex15p is required for peroxisomal localization of the GTPase Rho1p, which is thought to organize actin filaments on peroxisomes during proliferation [29]. The existence of a link between the AAA peroxins and matrix protein import has been proposed previously [30], but has remained elusive for many years. Recently, their functional role in peroxisomal protein import was discovered. The AAA peroxins are required for the dislocation of the cycling peroxisomal import receptors Pex5p and Pex20p from the peroxisomal membrane back to the cytosol in order to complete their receptor cycle [31–34]. The AAA peroxins function as dislocases for the ubiquitinated PTS1 (peroxisomal targeting signal 1) receptor Pex5p Import of folded proteins into peroxisomes occurs in a post-translational manner and depends on ATP. The soluble Seventh International Meeting on AAA Proteins Figure 1 Molecular organization of the AAA complex in S. cerevisiae The AAA peroxins Pex1p and Pex6p are composed of an NTD, a non-conserved AAA domain (D1) and a conserved AAA domain (D2). The AAA domains contain ATP-binding sites (A) and, with the exception of D1 of Pex6p, also ATP-hydrolysis sites (B). Pex1p and Pex6p form a heteromeric complex, and oligomerization requires the presence of the D1 domains and is stimulated by ATP binding to Pex1p D2. Recruitment of the AAA complex to peroxisomes occurs via binding of Pex6p NTD to Pex15p and requires ATP binding at Pex6p D1, while detachment from Pex15p needs ATP binding and hydrolysis at Pex6p D2. The peroxisomal AAA complex dynamically associates with the functional matrix protein-import machinery (importomer) and Pex4p (Ubc10p) is supposed to be required for the disconnection of the AAA complex from the importomer. PTS1 receptor Pex5p is the major signal-recognition factor of proteins destined for the peroxisomal matrix. The receptor cycle of Pex5p involves cargo recognition in the cytosol, docking of the receptor–cargo complex to the peroxisomal membrane, translocation of the receptor–cargo complex to the luminal side of the membrane, followed by release of the cargo into the matrix and retrotranslocation of the receptor back to the cytosol (Figure 2) [7]. Permeabilized cell systems of human fibroblasts provided the first evidence that Pex5p accumulated reversibly at the peroxisomal membrane under ATP-modulated conditions [30]. Detailed in vitro studies revealed that the binding and translocation of Pex5p itself is ATP-independent while the export of Pex5p back to the cytosol requires ATP [35]. The identity of the corresponding ATPase remained a matter of debate until in vitro systems in S. cerevisiae [34] and human fibroblast cells [32] identified Pex1p and Pex6p as the motor proteins of Pex5p export. Their function in this process requires the presence of their membrane-anchor proteins, Pex15p or Pex26p. The in vitro reconstitution of the complete Pex5p cycle revealed that ATP binding and hydrolysis at both Pex1p D2 and Pex6p D2 is needed for receptor dislocation [34]. Interestingly, the Walker B motif of Pex1p D2 seems to have no function in formation or targeting of the AAA complexes [11,12] and thus may be exclusively required for handling of the substrate. The binding and consumption of ATP is believed to induce conformational changes within the AAA peroxins that generate the driving force to pull the receptor out of the membrane by a mechanism possibly similar to the one of Cdc48p (p97/VCP) in ERAD (endoplasmic reticulum-associated degradation) [36]. The mechanism of substrate recognition by the AAA peroxins is not understood. Although Pex5p and the AAA proteins form a complex at the peroxisomal membrane [15,32,34], no direct interaction of the PTS1 receptor with either Pex1p or Pex6p has been reported. This interaction seems to be regulated or mediated by a third factor, which could represent an unknown adaptor protein of the AAA peroxins or post-translational modification of the substrate. It is well known that both processes play a central role in the function of Cdc48p (p97/VCP) [37,38], which is the closest evolutionary relative of Pex1p and Pex6p [39,40]. C The C 2008 Biochemical Society Authors Journal compilation 101 102 Biochemical Society Transactions (2008) Volume 36, part 1 Figure 2 Peroxisomal matrix protein import The AAA peroxins export the ubiquitinated PTS1 receptor back to the cytosol. The PTS1-recognition factor Pex5p binds newly synthesized PTS1-harbouring cargo proteins in the cytosol and ferries them to the docking complex (Pex13p, Pex14p, Pex17p) at the peroxisomal membrane. The receptor–cargo complex reaches the luminal side of the membrane, where the cargo is released in a process that possibly involves Pex8p. ATP-dependent retrotranslocation of the membrane-integrated Pex5p is mediated by the Pex15p-anchored AAA complex consisting of Pex1p and Pex6p. Pex5p can either be mono-ubiquitinated by the Pex22p-anchored Pex4p in order to be recycled (recycling pathway) or it can be polyubiquitinated by Ubc4p, Ubc5p and Ubc1p, resulting in proteasomal degradation (proteolytic pathway). Both pathways rely on ATP-dependent activation of ubiquitin by E1 and possibly require the RING (really interesting new gene) peroxins Pex2p, Pex10p and Pex12p as putative E3 enzymes. As a consequence, the question has to be addressed of how the AAA peroxins can distinguish Pex5p forms destined for dislocation from cargo-loaded Pex5p species destined for cargo translocation. A possible solution may arise from the crystal structure of Pex1p NTD, which displays similarities to the corresponding adaptor-binding domains of other AAA proteins [23]. Data from p97 and Ufd1 have identified a double-ψ β-barrel fold as a ubiquitin-binding domain with binding sites for both mono- and poly-ubiquitin [41]. Most interestingly, the PTS receptors Pex5p, Pex18p and Pex20p have been demonstrated to be ubiquitinated [31,42–44]. The PTS1 receptor Pex5p of S. cerevisiae is monoubiquitinated in wild-type cells [45], whereas it has been shown to be polyubiquitinated in mutants of the proteasome or cells affected in the AAA and Pex4p–Pex22p complexes of the peroxisomal protein-import machinery [42,43]. Polyubiquitination of Pex5p, requiring the ubiquitinconjugating enzymes Ubc4p and the partly redun C The C 2008 Biochemical Society Authors Journal compilation dant Ubc5p and Ubc1p, takes place exclusively at the peroxisomal membrane and marks the receptor for proteasomal degradation as part of a quality-control system [42,43,45]. Alternatively, Pex5p is the specific molecular target for mono-ubiquitination by Pex4p (Ubc10p) [33,46], which is essential for peroxisomal biogenesis [47] and is anchored via Pex22p to the peroxisomal membrane [48]. The functional role of ubiquitination in the dislocation process has been elucidated by in vitro export assays, revealing that mono-ubiquitination of Pex5p constitutes the export signal under physiological conditions, whereas polyubiquitination seems to provide an export signal for the release of dysfunctional PTS1 receptors from the membrane and proteasomal degradation as part of the quality-control pathway [33]. The direct mechanistic influence of this modification on the export reaction remains to be investigated. The AAA peroxins may interact directly or indirectly via putative Seventh International Meeting on AAA Proteins adaptors with the ubiquitin tag on Pex5p. Alternatively, the attachment of ubiquitin may induce local conformational changes within Pex5p to expose hidden binding sites. This mode of interaction is also discussed for Cdc48p (p97/VCP), which binds ubiquitin via adaptor complexes such as Ufd1/ Npl4 and via its N-terminal domain. This domain is capable of recognizing ubiquitin chains and also non-modified segments of its substrates [49,50]. Notably, the AAA complex displays significantly increased association with the importomer in PEX4-deficient cells, indicating that the ATPase cycles of Pex1p and Pex6p are coupled to the mono-ubiquitination-dependent receptor cycle of Pex5p (Figure 1) [15]. Conclusions Peroxisomes exhibit unique dynamics in their enzyme content and metabolic functions. The accompanied changes are accomplished by elaborate protein-transport machineries. The energy requirement for peroxisomal protein import is determined by the ATP-dependent dislocation of the import receptors, which probably represents the rate-limiting step. The energy is utilized by two enzyme activities: (i) monoubiquitination by Pex4p (recycling pathway) or polyubiquitination by Ubc4p (proteolytic pathway), as ubiquitin first has to be activated by E1; and (ii) ATP hydrolysis in the conserved AAA domains of Pex1p and Pex6p in order to pull the primed PTS receptor out of the membrane. These results bring together the previously disparate roles of Pex4p and the AAA peroxins in one concerted reaction sequence. For future research, it will be a challenge to elucidate how AAA-mediated receptor dislocation is mechanistically linked to the peroxisomal import of folded proteins. Note added in proof (received 13 December 2007) After submission of the present paper, an article appeared concerning the ubiquitination of mammalian Pex5p [51]. This study demonstrates that this modification is required for recycling and thus reveals that the mechanism of AAA peroxin function is highly censerved in evolution. We apologize to those scientists whose work could not be cited due to space limitations. We are grateful to Sigrid Wüthrich for technical assistance and Wolfgang Girzalsky and Marion Witt-Reinhardt for the reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB642, Er178/2-4), the FP6 European Union Project ‘Peroxisome’ (LSHG-CT-2004-512018) and by the Fonds der Chemischen Industrie. References 1 Erdmann, R., Wiebel, F.F., Flessau, A., Rytka, J., Beyer, A., Fröhlich, K.U. and Kunau, W.-H. (1991) PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64, 499–510 2 Kunau, W.-H., Beyer, A., Franken, T., Götte, K., Marzioch, M., Saidowsky, J., Skaletz-Rorowski, A. and Wiebel, F.F. (1993) Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie 75, 209–224 3 Neuwald, A.F., Aravind, L., Spouge, J.L. and Koonin, E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res. 9, 27–43 4 Erzberger, J.P. and Berger, J.M. (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 5 Hanson, P.I. and Whiteheart, S.W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 6 Voorn-Brouwer, T., van der Leij, I., Hemrika, W., Distel, B. and Tabak, H.F. (1993) Sequence of the PAS8 gene, the product of which is essential for biogenesis of peroxisomes in Saccharomyces cerevisiae. Biochem. Biophys. Acta 1216, 325–328 7 Erdmann, R. and Schliebs, W. (2005) Peroxisomal matrix protein import: the transient pore model. Nat. Rev. Mol. Cell Biol. 6, 738–742 8 Wanders, R.J. (2004) Metabolic and molecular basis of peroxisomal disorders: a review. Am. J. Med. Genet. 126A, 355–375 9 Reuber, B.E., Germain-Lee, E., Collins, C.S., Morrell, J.C., Ameritunga, R., Moser, H.W., Valle, D. and Gould, S.J. (1997) Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat. Genet. 17, 445–448 10 Geisbrecht, B.V., Collins, C.S., Reuber, B.E. and Gould, S.J. (1998) Disruption of a PEX1–PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc. Natl. Acad. Sci. U.S.A. 95, 8630–8635 11 Birschmann, I., Rosenkranz, K., Erdmann, R. and Kunau, W.-H. (2005) Structural and functional analysis of the interaction of the AAA-peroxins Pex1p and Pex6p. FEBS J. 272, 47–58 12 Tamura, S., Yasutake, S., Matsumoto, N. and Fujiki, Y. (2006) Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J. Biol. Chem. 281, 27693–27704 13 Fan, W. and Fujiki, Y. (2006) A temperature-sensitive CHO pex1 mutant with a novel mutation in the AAA Walker A1 motif. Biochem. Biophys. Res. Commun. 345, 1434–1439 14 Faber, K.N., Heyman, J.A. and Subramani, S. (1998) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 18, 936–943 15 Rosenkranz, K., Birschmann, I., Grunau, S., Girzalsky, W., Kunau, W.-H. and Erdmann, R. (2006) Functional association of the AAA-complex and the peroxisomal importomer. FEBS J. 273, 3804–3815 16 Tamura, S., Shimozawa, N., Suzuki, Y., Tsukamoto, T., Osumi, T. and Fujiki, Y. (1998) A cytoplasmic AAA family peroxin, Pex1p, interacts with Pex6p. Biochem. Biophys. Res. Commun. 245, 883–886 17 Weller, S., Cajigas, I., Morrell, J., Obie, C., Steel, G., Gould, S.J. and Valle, D. (2005) Alternative splicing suggests extended function of PEX26 in peroxisome biogenesis. Am. J. Hum. Genet. 76, 987–1007 18 Arlt, H., Tauer, R., Feldmann, H., Neupert, W. and Langer, T. (1996) The YTA10–12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85, 875–885 19 Glickman, M.H., Rubin, D.M., Fried, V.A. and Finley, D. (1998) The regulatory particle of the S. cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 20 Birschmann, I., Stroobants, A.K., Van Den Berg, M., Schäfer, A., Rosenkranz, K., Kunau, W.H. and Tabak, H.F. (2003) Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol. Biol. Cell 14, 2226–2236 21 Matsumoto, N., Tamura, S. and Fujiki, Y. (2003) The pathogenic peroxin Pex26p recruits the Pex1p–Pex6p AAA ATPase complexes to peroxisomes. Nat. Cell Biol. 5, 454–460 22 Furuki, S., Tamura, S., Matsumoto, N., Miyata, N., Moser, A., Moser, H.W. and Fujiki, Y. (2006) Mutations in the peroxin Pex26p responsible for peroxisome biogenesis disorders of complementation group 8 impair its stability, peroxisomal localization, and interaction with Pex1p–Pex6p complex. J. Biol. Chem. 281, 1317–1323 23 Shiozawa, K., Maita, N., Tomii, K., Seto, A., Goda, N., Akiyama, Y., Shimizu, T., Shirakawa, M. and Hiroaki, H. (2004) Structure of the N-terminal domain of PEX1 AAA-ATPase: characterization of a putative adaptor-binding domain. J. Biol. Chem. 279, 50060–50068 C The C 2008 Biochemical Society Authors Journal compilation 103 104 Biochemical Society Transactions (2008) Volume 36, part 1 24 Warner, D.R., Roberts, E.A., Greene, R.M. and Pisano, M.M. (2003) Identification of novel Smad binding proteins. Biochem. Biophys. Res. Commun. 312, 1185–1190 25 Seo, J.G., Lai, C.Y., Miceli, M.V. and Jazwinski, S.M. (2007) A novel role of peroxin PEX6: suppression of aging defects in mitochondria. Aging Cell 6, 405–413 26 Titorenko, V.I. and Rachubinski, R.A. (2000) Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J. Cell Biol. 150, 881–886 27 Titorenko, V.I. and Mullen, R.T. (2006) Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174, 11–17 28 Shiozawa, K., Goda, N., Shimizu, T., Mizuguchi, K., Kondo, N., Shimozawa, N., Shirakawa, M. and Hiroaki, H. (2006) The common phospholipid-binding activity of the N-terminal domains of PEX1 and VCP/p97. FEBS J. 273, 4959–4971 29 Marelli, M., Smith, J.J., Jung, S., Yi, E., Nesvizhskii, A.I., Christmas, R.H., Saleem, R.A., Tam, Y.Y., Fagarasanu, A., Goodlett, D.R. et al. (2004) Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 167, 1099–1112 30 Dodt, G. and Gould, S.J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763–1774 31 Leon, S., Zhang, L., McDonald, W.H., Yates, 3rd, J., Cregg, J.M. and Subramani, S. (2006) Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 172, 67–78 32 Miyata, N. and Fujiki, Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 33 Platta, H.W., El Magraoui, F., Schlee, D., Grunau, S., Girzalsky, W. and Erdmann, R. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204 34 Platta, H.W., Grunau, S., Rosenkranz, K., Girzalsky, W. and Erdmann, R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 35 Gouveia, A.M., Guimaraes, C.P., Oliveira, M.E., Reguenga, C., Sa-Miranda, C. and Azevedo, J.E. (2002) Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J. Biol. Chem. 278, 226–232 36 Pye, V.E., Dreveny, I., Briggs, L.C., Sands, C., Beuron, F., Zhang, X. and Freemont, P.S. (2006) Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 156, 12–28 37 Jentsch, S. and Rumpf, S. (2007) Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32, 6–11 C The C 2008 Biochemical Society Authors Journal compilation 38 Welchman, R.L., Gordon, C. and Mayer, R.J. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 39 Gabaldon, T., Snel, B., van Zimmeren, F., Hemrika, W., Tabak, H.F. and Huynen, M.A. (2006) Origin and evolution of the peroxisomal proteome. Biol. Direct 1, 1–14 40 Schluter, A., Fourcade, S., Ripp, R., Mandel, J.L., Poch, O. and Pujol, A. (2006) The evolutionary origin of peroxisomes: an ER–peroxisome connection. Mol. Biol. Evol. 23, 838–845 41 Park, S., Isaacson, R., Kim, H.T., Silver, P.A. and Wagner, G. (2005) Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13, 995–1005 42 Kiel, J.A., Emmrich, K., Meyer, H.E. and Kunau, W.H. (2005) Ubiquitination of the PTS1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280, 1921–1930 43 Platta, H.W., Girzalsky, W. and Erdmann, R. (2004) Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 384, 37–45 44 Purdue, P.E. and Lazarow, P.B. (2001) Pex18p is constitutively degraded during peroxisome biogenesis. J. Biol. Chem. 276, 47684–47689 45 Kragt, A., Voorn-Brouwer, T.M., Van den Berg, M. and Distel, B. (2005) The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 280, 7867–7874 46 Williams, C., van den Berg, M., Sprenger, R.R. and Distel, B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 47 Wiebel, F.F. and Kunau, W.-H. (1992) The PAS2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature 359, 73–76 48 Koller, A., Snyder, W.B., Faber, K.N., Wenzel, T.J., Rangell, L., Keller, G.A. and Subramani, S. (1999) Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J. Cell Biol. 146, 99–112 49 Thoms, S. (2002) Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett. 520, 107–110 50 Ye, Y., Meyer, H.H. and Rapoport, T.A. (2003) Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 51 Carvalho, A.F., Pinto, M.P., Grou, C.P., Alencastre, I.S., Fransen, M., Sá-Miranda, C. and Azevedo, J.F. (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 Received 28 August 2007 doi:10.1042/BST0360099 JBC Papers in Press. Published on June 10, 2011 as Manuscript M111.238600 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.M111.238600 Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery Mykhaylo O. Debelyy1, Harald W. Platta1,2, Delia Saffian1, Astrid Hensel1, Sven Thoms1,3, Helmut E. Meyer4, Bettina Warscheid4,5, Wolfgang Girzalsky1 and Ralf Erdmann1§ 1 Running head: A role for Ubp15p in Peroxisome Biogenesis Correspondence to: Dr. Ralf Erdmann, Institut für Physiologische Chemie, Ruhr-Universität Bochum, Universitätsstr. 150, D-44780 Bochum, Tel. 49-234-322-4943, Fax. 49234-321-4266, email. [email protected] § The peroxisomal matrix protein import is facilitated by cycling receptors shuttling between the cytosol and the peroxisomal membrane. One crucial step in this cycle is the ATP-dependent release of the receptors from the peroxisomal membrane. This step is facilitated by the peroxisomal AAAproteins Pex1p and Pex6p with ubiquitination of the receptor being the main signal for its export. Here we report that the AAA-complex contains dislocase as well as deubiquitinating activity. Ubp15p, an ubiquitin hydrolase, was identified as novel constituent of the complex. Ubp15p partially localizes to peroxisomes and is capable to cleave off ubiquitin-moieties from the PTS1-receptor Pex5p. Furthermore, Ubp15p-deficient cells are characterized by a stress related PTS1import defect. The results merge to a picture in which removal of ubiquitin of the PTS1-receptor Pex5p is a specific event and might represent a vital step in receptor recycling. Peroxisomes are organelles which carry out a wide variety of metabolic processes in eukaryotic organisms. As peroxisomes do not contain genetic material, their protein content is determined by the import of nuclear encoded proteins. Peroxisomes can multiply by division (1) or de novo by budding from the ER (2,3). Without exception, peroxisomal matrix proteins are synthesized on free ribosomes and are subsequently imported in a posttranslational manner (4,5). Like the sorting of proteins to other cellular compartments, protein targeting to peroxisomes depends on signal sequences. Peroxisomal matrix proteins contain a C-terminal type I peroxisomal targeting sequence (PTS1) or an N-terminal PTS2 (4). These PTSs are recognized by conserved receptors, Pex5p and Pex7p, respectively. Based on the concept of cycling receptors (6,7), the matrix protein import can be divided into four steps: 1) receptor-cargo recognition in the cytosol, 2) docking at the peroxisomal membrane, 3) cargo-translocation and release, and 4) receptor release from the membrane and recycling. With respect to the PTS1-receptor Pex5p, recent reports demonstrated that its dislocation from the peroxisomal membrane to the cytosol at the end of the receptor cycle is ATPdependent and catalyzed by the AAA-peroxins Pex1p and Pex6p (8,9). The main signal for the export process is the attachment of a monoubiquitin moiety or, alternatively, the anchoring of a polyubiquitin chain (10,11). While receptor monoubiquitination occurs on a conserved cysteine, polyubiquitin chains are Copyright 2011 by The American Society for Biochemistry and Molecular Biology, Inc. Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 Abteilung für Systembiochemie, Medizinische Fakultät der Ruhr-Universität Bochum, D-44780 Bochum, Germany 2 Current address: Department of Biochemistry, Institute for Cancer Research, The Norwegian Radium Hospital, Montebello, N-0310 Oslo, Norway 3 Current address: Universitätsmedizin Göttingen, Abteilung für Pädiatrie und Neuropädiatrie, Georg-August-Universität, D-37099 Göttingen, Germany 4 Medizinisches Proteom-Center, Ruhr-Universität Bochum, Universitätsstrasse 150, D-44780 Bochum, Germany 5 Current address: Faculty of Biology and BIOSS Centre for Biological Signalling Systems, University of Freiburg, 79104 Freiburg, Germany Here we report on the correlation of the ATPdependent export of Pex5p and ubiquitin cleavage. The AAA-complex of the peroxisomal protein import machinery turned out to possess export as well as deubiquitinating activity. Ubp15p was identified as novel constituent of the complex which binds to the first AAA domain of Pex6p (D1 domain). Ubp15p exhibits ubiquitin hydrolase activity and is capable to cleave off ubiquitin-moieties from the PTS1-receptor Pex5p. The function of Ubp15p in peroxisome biogenesis is supported by a stress related PTS1-import defect of ubp15Δ cells. A scenario evolves in which receptor deubiquitination might be functionally linked to its AAA-peroxin mediated export and represents an important step in the receptor cycle which makes Pex5p available for a new round of matrix protein import. EXPERIMENTAL PROCEDURES Yeast strains and culture conditions - The Saccharomyces cerevisiae strain UTL-7A (MATa, ura3-52, trp1, leu2-3/112) was used as wild-type strain for the generation of several isogenic deletion strains by the `short flanking homology` method as described previously (28). The resulting deletion strains were pex5Δ (29) ubp14Δ, ubp15Δ, ubp14Δ/ubp15Δ, doa4Δdoa 4Δ/ubp15Δ (this study). cl3-ABYS-86 (30) served as wild-type strain for isolation of His6Pex6p- and His6-GST-Ubp15p-complex. The yeast reporter strain L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3, URA3::lexA-lacZ) (31) was used for two-hybrid assays. Yeast media have been described previously (32). Inhibit of proteasomal degradation by the addition of MG132 to liquid cultures was performed according to (33) Plasmids and cloning strategies - Sequences of oligonucleotides available upon request. Twohybrid plasmids expressing Gal4p-fusions with Pex1p, Pex6p or variants thereof were described previously (34). For expression of His6-Ubp15p in bakers yeast, two overlapping PCRs were performed using genomic S. cerevisiae DNA as template. PCRI (primers RE1813/ RE1749) amplified the 5`- half of UBP15 (NTP-UBP15) introducing an NcoI site to the 5`end. PCRII (primers RE1746/RE1730) amplified the 3`-half of UBP15 (CTP-UBP15) introducing an XhoI site to the 3`-end. Both PCR-products were subcloned into EcoRV 2 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 attached to two lysine residues (10,12). In general, conjugation of ubiquitin to a target protein or to itself is regulated by the sequential activity of ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitinligating (E3) enzymes, and it typically results in the addition of an ubiquitin moiety either to the ε-amino group of a Lys residue or to the extreme amino terminus of a polypeptide (13). In a very few cases, including Pex5p, also attachment to a Cys residue has been reported (12,14). Whereas the addition of a single ubiquitin to a target protein can alter protein activity and localization, the formation of a diverse array of ubiquitin chains is implicated as targeting to the 26S proteasome (15). In line with these findings, polyubiquitination of Pex5p makes the receptor available for proteasomal degradation as part of a quality control system for the disposal of dysfunctional Pex5p (16-18). Modification of Pex5p by a single ubiquitin on a conserved Cys residue provides the signal for the AAAperoxin mediated release of the receptor from the peroxisomal membrane (10,11,19). This is of special importance as this ATP-dependent dislocation of the receptor is supposed to be responsible for the overall energy-requirement of the protein import cascade and thus might be mechanistically linked to the cargo translocation as proposed by the export-driven import model (20). The ubiquitination-cascade acting on Pex5p has been elucidated with the identification of Pex4p and the Ubc1p/Ubc4p/Ubc5p-family as responsible E2s (10,12,17,18,21). The peroxisomal RING-finger peroxins Pex2p, Pex10p and Pex12p have been identified as E3-enzymes responsible for the poly- and monoubiquitination of Pex5p (22,23). After export of the functional receptor to the cytosol, the ubiquitin-moiety has to be removed. This cleavage of ubiquitin from a substrate protein is generally carried out by ubiquitin hydrolases also known as deubiquitinating enzymes (DUBs) (24). S. cerevisiae contains genes coding for 18 DUBs (25,26). Recent in vitro data obtained from rat indicated that the monoUb moiety of Pex5p might be cleaved off in two different ways. A minor portion of the thioester-bound monoUb could be released in a non-enzymatic manner by a nucleophilic attack of glutathione while the major fraction of monoUb-Pex5p is deubiquitinylated enzymatically by a still to be identified ubiquitin hydrolase (27). Two-hybrid assay - The yeast reporter strain L40 was transformed with two-hybrid plasmids pPC86 and pPC97 (40) or derivates thereof and grown on synthetic medium lacking tryptophane and leucine for 3 days at 30 °C. Obtained double transformants were grown at 30 °C for 8 h in liquid synthetic medium. Lysates from these cells were prepared and subsequently subjected to ßgalactosidase assays as described by (41). Purification of Pex6p from S. cerevisiae cells Recombinant His-tagged Pex6p or Ubp15p were expressed in S. cerevisiae strain cl3ABYS-86 (30) transformed with pJK-5 or pYES263-UBP15, respectively. Galactosegrown cells were harvested, resuspended in lyses buffer (1.7 mM KH2PO4, 5.2 mM Na2HPO4, 300 mM NaCl, 1 mM DTT, 22.5 µg/ml DNase I) with protease inhibitors cocktail (8 µM antipain, 0.3 µM aprotinin, 1 µM bestatin, 10 µM chymostatin, 5 µM leupeptin, 1.5 µM pepstatin (Roche Diagnostics, Mannheim, Germany), 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 5 mM NaF). Cells were disrupted by glass bead lyses. Lysate was cleared by centrifugation and 0.22 µm filtration and loaded on Ni-Sepharose (GE Healthcare, Munich, Germany) columns equilibrated with washing buffer (1.7 mM KH2PO4, 5.2 mM Na2HPO4, 300 mM NaCl, 1 mM DTT, 40 mM imidazol).The column was washed until no more protein eluted. Pex6p was then eluted by a continuous imidazol gradient up to 500 mM imidazol in elution buffer (1.7 mM KH2PO4, 5.2 mM Na2HPO4, 300 mM NaCl, and 1 mM DTT, 500 mM imidazol). Fractions containing high protein concentration were combined and concentrated by VivaSpin concentrators (10,000 MWCO) (Sigma, Munich, Germany). Isolation of peroxisomes-Preparation of yeast spheroplasts, cell homogenization, preparation of post-nuclear supernatants and determination of the suborganellar localization of proteins were performed according to (42). Density gradient centrifugation was essentially performed as described (43), in particular, postnuclear supernatants (10 mg protein) were prepared and loaded onto preformed 2.25-24% (w/v) Optiprep (Iodixanol) gradients. Peroxisomes were separated from other organelles in a vertical rotor (Sorvall TV 860, 1.5 h at 48,000xg, 4°C). Fractions were collected from the bottom and subjected to enzyme and refractive index measurements as well as immunoblot analysis. Gel filtration of cell lysates and purified proteins - Analytical gel filtration was carried 3 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 digested vector pGEM®-T (Promega, Mannheim, Germany) resulting in vectors pGEM®-T-NTP-UBP15 and pGEM®-T-CTPUBP15, respectively. Next, the introduced fragments were cut out of the pGEM® vectors (pGEM®-T-NTP-UBP15, NcoI/BamHI pGEM®-T-CTP-UBP15, BamHI/XhoI) and cloned together into NcoI/XhoI sites of pYES263 (35) leading to pYES263-UBP15. For the expression of GST-fusions of Ubp15p in E. coli, the vector pGEX-4T-2 (GE Healthcare, Freiburg, Germany) was digested with BamHI followed by a Klenow-refill reaction and the subsequent cleavage with XhoI. The Ubp15p coding region was obtained by cleaving pYES263-UBP15 with NcoI, Klenow-based refilling and subsequent XhoI treatment. The UBP15 fragment was then cloned into equally treated pGEX-4T-2 resulting in pGEX-4T-2-UBP15. In order to introduce a C214A amino-acid residue substitution into Ubp15p, the QuickChange mutagenesis kit (Agilent Technologies, Waldbronn, Germany) was used combined with pGEX-4T2-UBP15 as template and primers RE2274/RE2275 for the reaction. GFP-Ubp15p expression plasmid pUG36UBP15 was constructed as follows. UBP15 was amplified by PCR using primers RE3196/RE3198 and plasmid pYES263UBP15 as template. The SpeI/SalI digested PCR product was cloned into SpeI/SalI site of pUG36 (36). To obtain N-terminal His6-TEV-tagged Pex6p under the control of the GAL1-promotor for expression in yeast, the coding region for the N-terminal half of Pex6p was amplified by PCR using primers KU1549/KU1550 and plasmid pMB34 (37) as template. In a second step, PEX6 was amplified by PCR (primers KU1339/KU698, plasmid pMB34) and cloned into NcoI/SpeI site of pYES2.1V5-His-TOPO (Invitrogen, Darmstadt, Germany) leading to vector pYQ6/1. Finally, the first PCR (Nterminal Pex6p half) was digested with PvuII/SacI and the fragment was introduced into PvuII/SacI digested vector pYQ6/1, leading to pJK-5. Plasmids for expression of PTS2-dsRed or high expression of Pex15p were described elsewhere (38,39). out on a SMART System (Amersham Pharmacia Biotech, Uppsala, Sweden) equipped with a Superose6 PC 3.2/30 column in running buffer (50 mM Tris/HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM βmercaptoethanol, 2 mM ATP). Samples were cleared by centrifugation (15 min, 20,000g) and aliquots of 50 µl purified protein were separated at 40 µl/min. Fractions of 80 µl were collected form 0.8 to 1.6 ml after injection. The column was calibrated using ferritin (440 kDa), aldolase (158 kDa), and BSA (66 kDa) as markers. Deubiquitination assay - Deubiquitinating activity of Ubp15p was analyzed according to (44). In detail, 1 µg of Ubp15p/Ubp15pC214A and 250 ng of appropriate polyUb(K63)-chains (Biomol, Loerrach, Germany) were diluted in reaction buffer (50 mM Tris-HCl, pH 7.4, and 300 mM NaCl) to a total volume of 30 µl. Reactions were incubated for 2 h at 37 °C. Before and after reaction, 15 µl of each sample were charged with 3x SDS sample buffer and boiled for 5 min for further analysis. Five µl of each reaction were loaded onto a 15 % trisglycin gel and subsequently subjected to immunoblot analysis. Protein Identification by Mass Spectrometry Proteins in polyacrylamide gels were visualized by Coomassie staining according to (45). Destaining of proteins, in-gel tryptic digestion and subsequent peptide extraction was performed as described (46). Peptide samples were separated by online reversedphase nano-HPLC using the Dionex LC Miscellaneous - Immunopurification of ProtAtagged Pex1p/Pex6p-complexes from yeast cells using IgG-Sepharose was described in (47). Immunoprecipitation of denatured proteins was carried out according to (22). Membranes containing monoubiquitinylated Pex5p were prepared according to (22) and incubated with purified yeast AAA-complex according to (8). Recombinant GST-fusionproteins were expressed in E. coli BL21 (DE3) according to manufactures protocols (GE Healthcare, Freiburg, Germany). Immunoreactive complexes were visualized using antirabbit or anti-mouse IgG-coupled horseradish peroxidase in combination with the ECL™ system from Amersham Pharmacia Biotech (Uppsala, Sweden). Alternatively, primary antibody was detected with a IRDye 800CW goat anti-rabbit IgG secondary antibody (LICOR Bioscience, Bad Homburg, Germany) followed by a detection using the “Infrarot Imaging System“ (LI-COR Bioscience, Bad Homburg, Germany). Polyclonal rabbit antibodies were raised against Pex5p (48), Pex13p (29), and ubiquitin (Sigma, Munich, Germany). Monoclonal mouse antibodies were raised against GST (Sigma, Munich, Germany) and ubiquitin (clone FK2, Biomol, Hamburg, Germany). GFP- and dsRed-tagged proteins were monitored by life cell imaging with a Zeiss Axioplan 2 fluorescence microscope and AxioVision 4.8 software (Zeiss, Jena, Germany). Electron transmission microscopy, spheroplasting of yeast cells, homogenization and differential centrifugation at 25,000 x g of homogenates were performed as described previously (8,42,49). RESULTS The peroxisomal AAA-complex exhibits deubiquitinating activity Dislocation of the PTS1 receptor Pex5p from the peroxisomal membrane to the cytosol depends on the peroxisomal AAA-proteins Pex1p and Pex6p (8,9) and ubiquitination of Pex5p is a prerequisite for this process (10). 4 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 In vivo ubiquitination assays - Oleate-induced yeast cells were harvested, washed twice and resuspended in lysis-buffer (0.2 M HEPES, 1 M potassium acetate and 50 mM magnesium acetate, pH 7.5) and protease inhibitors cocktail (see above), 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 5 mM NaF). To accumulate monoubiquitinated Pex5p from wild-type cells, 20 mM NEM (Sigma, Munich, Germany) was added. Cells were disrupted by glass bead lysis and centrifuged at 1,500xg (Eppendorf rotor A-481) for 10 min. Supernatants were normalized for protein and volume, and membranes were sedimented by centrifugation at 100,000xg, (30 min, Sorvall AH650 rotor) followed by trichloroacetic acid precipitation and sample preparation (18). Packings HPLC systems (Dionex LC Packings, Idstein, Germany). Electrospray ionization tandem mass spectrometry (ESI-MS/MS) on a Bruker Daltonics HCTplus ion trap instrument (Bremen, Germany) and subsequent protein identification by bioinformatics using the yeast NCBI database was performed as described (46). Ubp15p is associated with the AAA-complex The data described above indicate that the isolated AAA-complex contains export- and deubiquitinating activity also in the absence of the cytosol. Thus, the suspected additional factor is supposed to be part of the yeast AAAcomplex. To identify the unknown factor, we isolated the cytosolic AAA-complex with Pex6p as overexpressed bait protein. For this purpose, a plasmid encoding N-terminal His6tagged Pex6p under the inducible GAL1promotor was transformed into the protease deficient yeast strain cl3-ABYS-86 (30). Transformants were precultured on glucose rich media and expression of the tagged Pex6p was induced by shifting to galactose media. His-Pex6p was isolated by affinity chromatography on NiNTA and analyzed by SDS-PAGE followed by silver stain. Two dominant protein bands were visible (Fig. 2A) which have been excised and analysed by mass spectrometry. The fast migrating protein was identified as the bait protein Pex6p. The band with an approximate size of 140kDa consisted of three proteins, Clu1p, Ubp15p and Ecm21p. Clu1p is a subunit of translation initiation factor eIF3 that functions in AUG scanning in translation which is also required to maintain the morphology of mitochondria (51,52). Ubp15p is an ubiquitin-specific processing protease (53). Ecm21p is an arrestin-related protein which acts as an adaptor in ubiquitin ligation (54). As a second approach to identify AAA-peroxin associated proteins, we genomically tagged Pex1p with Protein A, isolated the complex as previously described (8), separated proteins of the isolated complex by SDS-PAGE and subjected selected protein bands to mass spectrometric analysis. The band marked in Fig. 2B contained Ubp15p, which indicated its association with the AAAcomplex and moved the protein into the focus of our interest. To validate the Pex6p-Ubp15pinteraction, the complex isolation was performed vice versa using Ubp15p as bait. His6-GST-tagged Ubp15p was expressed in wild-type cl3-ABYS-86 strain, isolated by immuno-purification and the constituents of the complex were analyzed by immunoblotting. Pex6p was identified as a component of the Ubp15p-complex (Fig. 2C) and a minor portion of the PTS1-receptor Pex5p also co-eluted with the Ubp15pcomplex. The soluble fructose 1,6bisphosphatase (Fbp1p, (55)) was not retained by the chromatography, an indication for the specificity of the isolation procedure (Fig. 2C). A portion of the ubiquitin hydrolase Ubp15p localizes to peroxisomes Our results demonstrate that yeast Ubp15p possesses the ability to interact with Pex6p. As Pex6p is localized in the cytosol and at peroxisomes, the subcellular localization of Ubp15p was analyzed under peroxisomeinducing conditions. To this end, a cell free homogenate of oleic acid-induced wild-type cells expressing a genomically tagged UBP15 gene coding for a Ubp15p-protein A fusion protein (Ubp15p-TEV-ProtA) was prepared and organelles were separated by density gradient centrifugation (Fig. 3A). The presence of organelle marker proteins in fractions was assayed either by determination of enzyme activities or by immunoblotting. As indicated by the segregation behaviour of the peroxisomal membrane marker Pex13p and activity measurements of the peroxisomal 5 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 The main signal for the export process is the attachment of a monoubiquitin moiety (10,11). In order to gain more insight into the principal export mechanism of monoubiquitinated Pex5p, membranes were prepared from wildtype cells in the presence of NEM (N-ethylmaleimid), which is a suitable inhibitor of deubiquitinating enzymes (DUBs) (50) and results in the accumulation of monoubiquitinated Pex5p at the peroxisomal membrane (Kragt et al., 2005; Platta et al., 2007). As NEM proved to be a competent inhibitor of the export machinery, membranes were washed extensively and NEM was avoided upon purification of the AAAcomplexes. The membranes containing monoubiquitinated Pex5p were incubated with buffer alone or buffer containing purified cytosolic AAA-complex of S. cerevisiae in the presence of an ATP-regenerating system (ARS, (8)) for 30 min at 37 °C. Interestingly, the presence of the AAA-complex did result in the disappearance of the monoUb-Pex5p (Fig. 1, lanes 1 and 2) indicating that the peroxisomal AAA-complex does not only harbor the known dislocase activity of Pex1p/Pex6p but is also capable to facilitate receptor deubiquitination in addition. This assumption is supported by the result that incubation of the AAA-complex with DUBinhibitors NEM (Fig. 1, lane 3) or ubiquitinaldehyde (Fig. 1, lane 4) prior the assay blocks Pex5p deubiquitination. Ubp15p interacts with the first AAA-domain of Pex6p In order to analyze the Ubp15p-interaction with the peroxisomal AAA-complex in more detail, we applied the yeast two-hybrid system. Plasmids expressing full-length Pex1p, Pex6p or ubiquitin fused to the Gal4p activation domain or the Gal4p-DNA-binding domain were transformed in the S. cerevisiae strain L40, and reporter gene expression was analyzed by assaying β-galactosidase activity. In line with previous findings (57), coexpression of Ubp15p with ubiquitin leads to significant reporter-gene activity as judged by determined ß-galactosidase activity, which indicated the known Ubp15p-ubiquitin interaction (Fig. 4). The enzyme activities differed in dependence of whether Ubp15p was fused to the DNA-binding or trans-activation domain of Gal4p, however, in case of an interaction the enzyme activity was significantly higher than the controls of the empty vector versus bait plasmids. Comparison of the different assays revealed that Pex6p interacts with Ubp15p while the monitored βgalactosidase activity was only slightly above the control level when Pex1p was tested for interaction with Ubp15p. To determine the Pex6p-region that contributes to the Ubp15p interaction, we analyzed the interaction of Ubp15p with the N-terminal region (N, aa1– 428), the first AAA-cassette (D1, aa421–716) and the second AAA-cassette (D2, aa704– 1030) of Pex6p and combinations thereof. As shown in Fig. 3, neither the N-domain nor the second AAA-domain is capable to interact with Ubp15p. In contrast, the first AAAdomain of Pex6p alone or fused to the Ndomain led to ß-galactosidase activity in the same range as observe with full-length Pex6p. Thus, the first AAA-domain of Pex6p is involved in the interaction with Pex1p (34,58) as well as with Ubp15p. Ubp15p facilitates deubiquitination of Pex5p in vitro UBPs represent a subclass of the deubiquitinating enzymes (DUB) comprising 18 putative members in S. cerevisiae, including Ubp15p (53). The UBP-family is highly divergent, but all members contain several short consensus sequences, the Cys- and the His- boxes that are likely to form a part of the active site (59). Within Ubp15p, the Cys-box covers the amino acids 206 to 223, whereas the His-box is localized between amino acids 449 and 533 (60). Sequence alignment of Ubp15p with other UBPs indicated that Cys214 of Ubp15p most likely represents an amino acid residue which is crucial for the deubiquitinating activity (60). Accordingly, a Cys214 to Ala substitution was introduced into the full-length protein by site directed mutagenesis and recombinant wild-type or mutant Ubp15p (Ubp15pC214A) fused to GST were expressed in E. coli and isolated by affinity chromatography. The tag was removed by thrombin cleavage. To demonstrate that recombinant Ubp15p exhibits deubiquitinating activity and is thus biologically active, in vitro ubiquitin-cleavage assays were performed. To 6 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 catalase, peroxisomes migrated to the bottom fractions and showed a clear peak in fractions 3, clearly separated from the mitochondrial marker (porin). In line with the reported cytosolic localization (56), the majority of Ubp15p remained in the top gradient fractions. However, a significant portion of Ubp15p is detected at a higher density, co-localizing with peroxisomal marker proteins (Fig. 3A, lane 3). To support this finding, we monitored localization of GFP-tagged Ubp15p by fluorescence microscopy. GFP-Ubp15p was co-expressed with the synthetic peroxisomal marker protein PTS2-dsRed in wild-type cells. PTS2-dsRed exhibits punctate fluorescence pattern, typically for a peroxisomal localization (Fig. 3B, (38)). In line with our results obtained by cell fractionation (Fig. 3A), GFPUbp15p was predominantly localized to the cytosol which leads to overall cellular fluorescence (Fig. 3B). However, a portion of GFP-Ubp15p co-stained with PTS2-dsRed positive structures demonstrating its peroxisomal localization. Next we tried to increase the amount of peroxisomal Ubp15p by overexpression of Pex15p. The overexpression of this peroxisomal membrane protein leads to an increased recruitment of Pex6p to peroxisomes (37). We assumed that the consequence thereof should be an increased amount of Ubp15p bound to the peroxisomal membrane, as it is a binding partner of the Pex6p-complex. Indeed, upon Pex15poverexpression only a small portion of GFPUbp15p was found cytosolic, whereas the major fraction was found co-localized with peroxisomal marker PTS2-dsRed (Fig. 3B). Taken together, the localization studies indicate that a portion of Ubp15p is associated with peroxisomes. together, the data demonstrate that recombinant Ubp15p exhibits ubiquitin hydrolase activity and facilitates deubiquitination of mono- and poyUb-Pex5p. Clustered peroxisomes in ubp15∆ cells Ubp15p is a cytosolic protein, which is associated with the yeast AAA-complex. Pex1p and Pex6p are both required for peroxisomal matrix protein import, leading to the question whether also Ubp15p contributes to peroxisomal function in vivo. To address this question, growth test were performed on plates containing oleic acid as sole carbon source, which will support cell growth only if peroxisomal ß-oxidation is functional, which requires an intact organelle biogenesis. In contrast to wild-type, PEX5-deficient cells are unable to grow on this medium, which is in accordance with the literature (62) and typical for peroxisomal mutant strains of S. cerevisiae (42). Cells deficient in Ubp15p did not exhibit a growth defect on oleic acid medium (Fig. 6A). As a partial defect in peroxisome biogenesis does not inevitably lead to a complete destruction of peroxisome function, we analyzed the matrix protein import in wildtype and mutants in more detail. To this end, the subcellular localization of GFP fused to the peroxisomal targeting signal 1 (GFP-PTS1) as marker for the Pex5p dependent import, and PTS2-dsRed, an artificial substrate for the Pex7p dependent matrix protein import were monitored by live cell imaging. Fluorescence microscopy inspection of oleic acid-induced wild-type cells revealed a punctuate staining pattern for both marker proteins, typical for a peroxisomal labeling (Fig. 6B). Mutant pex5Δ cells that are affected in peroxisomal protein import of PTS1-proteins (62) exhibited a cytosolic fluorescence pattern for GFP-PTS1 as typical for these cells. In contrast, the PTS2pathway is not affected in pex5Δ cells which results in a punctuate staining pattern for PTS2-dsRed. The fluorescence microscopy pattern observed for the ubp15∆ strain was similar to the one visible in the wild-type strain (Fig. 6B), suggesting that ubp15Δ cells are still able to import both, PTS1- and PTS2containing peroxisomal matrix proteins. Interestingly, in contrast to wild-type peroxisomes, which are well separated, peroxisomes of ubp15Δ cells appeared to form clusters (Fig. 6B). This observation was corroborated by electron microscopic 7 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 this end, the isolated proteins were incubated with Ub-chains and the reaction was stopped after zero (control) or 120 min by adding SDSsample buffer and subsequent boiling. Cleavage of the Ub-chain was monitored by immunoblot analysis with an antiserum against ubiquitin. Incubation of the Ub-chain with wild-type Ubp15p resulted in a decrease of higher molecular Ub-species and accumulation of monoUb as cleavage product (Fig. 5A, lane 2). When the assay was performed with mutated Ubp15p, no difference between the control sample and the sample incubated for 120 min (Fig. 5A, lanes 3 and 4) was observed. Thus, our data are clear in that Ubp15p acts as ubiquitin hydrolase on Ubchains and that an enzyme harboring the Cys214Ala replacement is enzymatically inactive. This finding is not due to a dramatic influence of the mutation on the structure of the protein as both wild-type as well as mutated Ubp15p exhibit same behavior when analyzed by size exclusion chromatography (data not shown). Pex5p can be monoubiquitinated (21) or polyubiquitinated (17,18). In Fig. 1, we showed that the AAA-peroxin complex harbors deubiquitinating activity. Now, we addressed the question whether mono- or polyubiquitinated Pex5p can function as molecular target for deubiquitination by Ubp15p. To this end, we prepared membranes from wild-type cells in the presence of NEM which results in the accumulation of monoUbPex5p. These membranes were incubated with recombinant Ubp15p, followed by co-immunoisolation of Pex5p. MonoUb-Pex5p was visible when the membranes were incubated with buffer alone but disappeared upon incubation with Ubp15p (Fig. 5B). Next, we assayed whether Ubp15p also acts on polyubiquitinated Pex5p. We isolated whole cell membranes from a pex1Δpex6Δ strain. These membranes show accumulation of polyUb-Pex5p species (17,18), Fig. 5C, lane 1). Incubation of these membranes with recombinant Ubp15p resulted in disappearance of modified Pex5p, indicating that Ubp15p can also cleave off ubiquitin from polyUb-Pex5p (Fig. 5C, lane 4). In line with this finding, no cleavage of Pex5p was observed when Ubp15p activity was blocked by NEM or Ub-aldehyde (Fig. 5C, lanes 2 and 3). Ub-aldehyde inhibits ubiquitin hydrolases by the formation of an extremely tight complex, in which the inhibitor is bound to the active site of DUBs (61). Taken of ubiquitin chains (59). Doa4p is required for turnover of the PTS2-co-receptor Pex18p, which also is ubiquitinated (63). Interestingly, Doa4p interacts with Ubp15p as well as Ubp14p (64,65). Thus, Ubp15p might be part of a ternary complex of ubiquitin hydrolases with overlapping functions. For this reason, we analyzed single mutants of UBP15, UBP14 and DOA4 as well as combination thereof for their capacity to import GFP-PTS1. As judged by fluorescence microscopy, neither the singlenor the double deletion strains exhibited an import deficiency for GFP-PTS1 (Fig. 8A, left panel). In all strains tested, the GFP-SKL exhibited a clear punctuate staining, demonstrating its localization in the peroxisomal matrix. It is well known that the function of redundant protein sometimes becomes essential when cells are under stress (66). To test for this possibility, we examined oleic acid induced wild-type and mutant cells for matrix protein import upon oxidative-stress conditions (0.2 mM H2O2 (67)). Under this condition, neither wild-type nor ubp14Δ cells showed an import defect for GFP-SKL as indicated by the clear punctuate staining with no background labeling (Fig. 8A, right panel). In contrast, doa4Δ and ubp15Δ cells showed a punctuate staining of peroxisomal matrix marker GFPSKL but also a background labeling indicative for a partial mislocalization of the marker protein to the cytosol. This finding indicated that these mutants exhibit a partial peroxisomal protein import defect upon oxidative-stress. In this respect, it is worth to note that expression of Ubp15p and Dao4p but not of Ubp14p is induced by oleic acid (68). Moreover, induction of Doa4p and Ubp15p is also induced upon oxidative-stress by H2O2 (69). Our data indicate that deficiency in both, Ubp15p or Doa4p, affects proper peroxisomal protein import under oxidative-stress condition. To support this observation of an impaired peroxisomal function under oxidative-stress, we monitored the growth behaviour of UBP15affected cells in comparison with wild-type and doa4Δ-cells. Cells were grown on either glucose or oleic acid as sole carbon source in the absence or presence of 0.2 mM H2O2. As judged by optical density measurements, wildtype as well as ubp15Δ cells exhibit similar growth rates when glucose served as energy source (Fig. 8B, lower panel). When H2O2 was added to the media, growth rates of these 8 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 inspection of wild-type and mutant cells (Fig. 6C). ubp15Δ-cells exhibit lower steady state concentration of Pex5p but higher rate of ubiquitinated Pex5p. To investigate the consequence of a deletion of UBP15 on turnover of Pex5p, we estimated Pex5p steady state concentration in wild-type and ubp15Δ-cells. Whole cell lysates of oleicacid induced wild-type and ubp15Δ-cells were subjected to immunoblot analysis with mitochondrial porin as loading control. Although porin concentration was same in both strains, Pex5p level differed (Figure 7A, left panel). For quantification of the observed difference, the signal intensity was analyzed by densitometry. It turned out that the Pex5p amount in ubp15Δ cells is reduced to approximately half of the wild-type level (Figure 7A, right panel). Next we analyzed whether the lower amount of Pex5p in ubp15Δ-cells is accompanied by a higher ubiquitination rate. To this end, we monitored receptor ubiquitination in cells treated with the MG132, which inhibits proteasomal protein degradation and leads to accumulation of ubiquitinated Pex5p (22). Accordingly, , ubiquitinated Pex5p was visible in both MG132-treated wild-type and ubp15Δ-cells (Figure 7B, left panel). However, while the level of unmodified Pex5p in ubp15Δ cells was half of wild-type level as described before (Figure 7B), ubp15Δ exhibits three times more polyubiquitinated Pex5p than the wild-type strain. Taken together, our results indicate a higher polyubiquitination rate of Pex5p in stains lacking Ubp15p which most likely causes a reduced steady state concentration of the PTS1-receptor. ubp15Δ-cells exhibit oxidative-stress related import deficiencies and growth on oleic acid None of the genes encoding ubiquitin hydrolases are essential for viability, suggesting that many of these enzymes have overlapping functions (53). As ubp15Δ exhibits normal growth and matrix protein import under oleic acid conditions, we suspected such a redundancy and tested double-deletion strains for growth on oleic acid medium and peroxisomal protein import. Cells lacking Doa4p are characterized by decreased free ubiquitin levels and these mutants display a strongly reduced turnover of several proteins that are targeted to degradation via ubiquitination. In line with this observation, Doa4p was shown the be involved in cleavage DISCUSSION The AAA-complex of Pex1p and Pex6p is responsible for the release of the ubiquitinated PTS1-receptor Pex5p from the peroxisomal membrane, which has been regarded as the final step of the peroxisomal protein import cascade. In this work, we show that the AAAcomplex is also responsible for receptor deubiquitination, which is supposed to be an important step in receptor recycling. We have identified the corresponding ubiquitin hydrolase Ubp15p as a novel factor that accompanies the AAA-complex in peroxisomal protein import. PolyUb-Pex5p (17,18) as well as monoUbPex5p (21,27) are solely found at the peroxisomal membrane fraction in wild-type yeast and rat liver cells, indicating that Pex5p ubiquitination exclusively takes place at the peroxisomal membrane. Interestingly, exported Pex5p appears to be unmodified, indicating that the Ub-moiety is removed during or directly after receptor export (10,11,21). However, published data on the deubiquitination of Pex5p so far have focused on in vitro assays with mammalian Pex5p. Soluble monoUb-Pex5p is formed when the in vitro export reaction is performed in presence of DUB inhibitors (11,27). Accordingly, it was concluded that deubiquitination of Pex5p occurs predominantly in the cytosol after release from the membrane. It also was suggested that a small fraction of the dislocated Ub-Pex5p in vitro can already be deubiquitinated by reducing reagents like glutathione, while most of the Ub-Pex5p is deubiquitinated via an enzymatic pathway (27). However, cleavage of the Ub-moiety from mammalian Pex5p was thought to be catalyzed by an unspecific reaction that could be carried out by any DUB in the cytosol or may even function via a non-enzymatic reaction. Our data indicate that deubiquitination of yeast Pex5p represents a specific and important event for the optimal functionality of the export machinery. With Ubp15p, we have identified a deubiquitinating enzyme that is dedicated for this deubiquitination event in baker`s yeast. The deubiquitinating activity found to be associated with the endogenous AAA-complex was the first indication for the presence of such an enzyme. Mass spectrometry analysis of the AAA-complex derived from endogenous proteins as well as overexpressed Pex6p revealed a stable association of Ubp15p. The interaction with Pex6p was confirmed by yeast two-hybrid analysis and the interaction site could be mapped to the D1 domain of Pex6p. While the evolutionarily related AAA-protein Cdc48p(p97/VCP) utilizes several co-factors (71), Ubp15p is only the second known cofactor that accompanies the function of Pex6p, with its membrane-anchor Pex15p (Pex26p in mammals) being the first one (37). Pex6p acts in concert with Pex1p as dislocase complex for the ubiquitinated Pex5p in order to facilitate the export of the PTS1-receptor back to the cytosol (8,9). This leads to the intriguing question, how the activity of the deubiquitinating enzyme Ubp15p is coordinated with the Ub-dependent dislocation of Pex5p from the membrane and release into the cytosol. The finding that the deletion of UBP15 does not result in a complete peroxisomal biogenesis defect, can either be explained by the model, that deubiquitination has only modulating activity or it may indicate the existence of additional factors which may accompany the AAA-complex in its function. This situation could well be explained by redundant DUBs acting on Ub-Pex5p. Possible candidates are the known Ubp15p-binding partners Ubp14p and Doa4p (Ubp4p) (64,65). However, the characterization of the single deletion strains suggested that these two DUBs do not have a peroxisome-specific function 9 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 strains were nearly the same as without oxidative-stress. In contrast, doa4Δ cells showed the lowest growth rate of monitored strains already without H2O2, but growth was significantly delayed in the presence of H2O2. When cells were cultured on oleic acid medium, growth of doa4Δ cells was drastically reduced (Fig. 8B, lower panel), which is in line with known pleiotropic effects of the deletion of this protein as Doa4p is involved in many Ub-dependent processes in the cell (70). The growth rate of wild-type and ubp15Δ cells was similar on glucose medium also under oxidative-stress conditions. Both strains also grew equally well on oleic acid medium without H2O2. However, oxidative-stress affected growth on oleic acid medium of both wild-type and ubp15Δ cells but while the wildtype still did grow reasonable well, growth of the ubp15Δ cells was severely affected (Fig. 8B, lower panel). Taken together, analysis of the mutant phenotypes disclosed a peroxisome- and stress-related defect of ubp15Δ cells. Although Ubp15p is not essential for peroxisomal biogenesis under normal conditions, its regulative function gains significantly more weight when the cells are stressed with H2O2 and require an efficient import of matrix proteins into peroxisomes. Thus, the findings that 1) Ubp15p is stably associated with the export machinery by interacting with Pex6p, 2) the fact that a small portion of the protein is associated with peroxisomes, and 3) the partial protein import defect for PTS1 proteins observed in ubp15Δ cells upon oxidative stress suggest that the deubiquitination, at least in bakers yeast, is not an unspecific event that takes place at any location in the cytosol, as suggested by the mammalian study (27), but supports the notion that the detachment of the Ub-moiety is a regulated event. Ubiquitination of the receptor is a precondition for its export (10,11). In this respect, it is likely that the Pex1p/Pex6p-complex recognizes the Ub-moiety. This, however, still needs to be shown. The in vitro data demonstrate that the exported import receptor is deubiquitinated. This reflects the in vivo situation which is clear in that the cytosolic receptor is not ubiquitinated. Thus, the accumulating evidence indicates that the ubiquitin moiety is cleaved off from the import receptor during or shortly after export. There are several possible advantages to favor a peroxisome-associated deubiquitination of Ub-Pex5p. This could protect Pex5p from unspecific ubiquitination by detaching Ub-moieties from lysine residues or preventing the formation of a poly-ubiquitin chain at the crucial cysteine residue dedicated to mono-ubiquitination. This function would ensure an optimal protection and presentation of monoUb-Pex5p to the export machinery. Another possible explanation might be that the deubiquitination step may trigger the efficient release of Pex5p from the export-machinery by cleavage of the complex-bound Ub-moiety. Furthermore, this mechanism could prevent the monoUb-Pex5p to be recognized by the proteasome system to ensure an efficient recycling of the receptor for new matrix protein import cycles. The finding that both ubiquitinating and deubiquitinating activities are required for the transport of proteins from a membrane to the cytosol finds an examples in the ERAD pathway. The AAA-type ATPase p97(Cdc48/VCP) is evolutionary related to the peroxisomal AAA-proteins Pex1p and Pex6p (76). Most interestingly, among the growing number of known co-factors and adaptor proteins that p97 utilizes to carry out its different functions are also several deubiquitinating enzymes (71). The mammalian deubiquitinating enzymes YOD1 and Ataxin-3 are p97-associated proteins and function in the ERAD pathway (77-79). Most of the published literature defines both DUBs as a positive regulator of the p97-driven dislocation of the ERAD-substrates, most likely by editing the poly-Ub chains on the 10 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 similar to Ubp15p. The single deletion strain of Ubp14p had no significant effect on peroxisome morphology or cargo import, both under oleate as well as under H2O2 stress conditions. Previous studies have suggested a role for Ubp14p in the disassembly of unanchored polyubiquitin chains (64). The deletion of Doa4p had an effect on the efficiency of peroxisomal cargo import. However, it has to be taken into account that the deletion of Doa4p is known to result in pleiotropic effects on many Ub-dependent processes in the cell, as Doa4p influences the homeostasis of free ubiquitin (70). Possibly related to this function, DOA4 is a stress regulated gene, giving an alternative explanation for the oleate induction reported by (68). Thus, although we cannot fully exclude that Doa4p exhibits a peroxisomerelated overlapping function with Ubp15p, the partial import defect observed for the doa4Δ strain might well be explained by the pleiotropic phenotype of this mutant. The observation that ubp15Δ cells contain more clustered peroxisomes than wild-type cells is puzzling. Earlier work correlated a reduced level of imported matrix proteins such as catalase and the occurrence of clustered peroxisomes (72). Slowing of Pex5p cycling is most certainly associated with reduced import rates. Interestingly, induction of oxidative stress by treating cells with hydrogen peroxide causes Pex5p to amass on the organelle membrane and significantly reduces PTS1 protein import (73-75). As our data are clear in that Ubp15p can deubiquitinate Pex5p and as the ubiquitination status of the PTS1-receptor directly influences its cycling (10,11), it is conceivable that the deletion of Ubp15p influences the import process of PTS1-proteins like catalase and thus possibly also morphology and clustering of peroxisomes. substrates themselves in order to ensure the best fit to downstream Ub-receptor proteins. Ubp15p acts in concert with the AAA-peroxins in the matrix protein import cycle of the PTS1receptor. Pex5p deubiquitination occurs as a highly specific event in yeast and removal of ubiquitin of the PTS1-receptor Pex5p turns out to be a vital step in the receptor cycle in its own right. Thus, removal of the ubiquitin seems to complete the receptor cycle of Pex5p in order to make the receptor available for another round of matrix protein import. ACKNOWLEDGMENTS We are grateful to Ulrike Freimann for technical assistance and to Wolfgang Schliebs for the reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB642). H.W.P was supported by an EMBO Long term Fellowship. 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. Motley, A. M., and Hettema, E. H. (2007) J. Cell Biol. 178(3), 399-410 Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005) Cell 122(1), 89-95 Lam, S. K., Yoda, N., and Schekman, R. (2010) Proceedings of the National Academy of Sciences of the United States of America Doi: 10.1073/pnas.1013397107 Schrader, M., and Fahimi, H. D. (2008) Histochem. Cell. Biol. 129(4), 421-440 Ma, C., and Subramani, S. (2009) IUBMB life 61(7), 713-722 Dodt, G., and Gould, S. J. (1996) J. Cell Biol. 135, 1763-1774 Marzioch, M., Erdmann, R., Veenhuis, M., and Kunau, W.-H. (1994) EMBO J. 13(30), 4908-4918 Platta, H. W., Grunau, S., Rosenkranz, K., Girzalsky, W., and Erdmann, R. (2005) Nat. Cell Biol. 7(8), 817-822 Miyata, N., and Fujiki, Y. (2005) Mol. Cell. Biol. 25(24), 10822-10832 Platta, H. W., El Magraoui, F., Schlee, D., Grunau, S., Girzalsky, W., and Erdmann, R. (2007) J. Cell Biol. 177(2), 197-204. Carvalho, A. F., Pinto, M. P., Grou, C. P., Alencastre, I. S., Fransen, M., Sá-Miranda, C., and Azevedo, J. E. (2007) J. Biol. Chem. 282(43), 31267-31272 Williams, C., van den Berg, M., Sprenger, R. R., and Distel, B. (2007) J. Biol. Chem. 282(31), 22534-22543 Hicke, L., Schubert, H. L., and Hill, C. P. (2005) Nat. Rev. Mol. Cell Biol. 6(8), 610621 Cadwell, K., and Coscoy, L. (2005) Science 309(5731), 127-130 Dikic, I., Wakatsuki, S., and Walters, K. J. (2009) Nat. Rev. Mol. Cell Biol. 10(10), 659-671 Erdmann, R., and Schliebs, W. (2005) Nat. Rev. Mol. Cell Biol. 6(9), 738-742 Kiel, J. A., Emmrich, K., Meyer, H. E., and Kunau, W. H. (2005) J. Biol. Chem. 280(3), 1921-1930 Platta, H. W., Girzalsky, W., and Erdmann, R. (2004) Biochem. J. 384, 37-45 Grou, C. P., Carvalho, A. F., Pinto, M. P., Wiese, S., Piechura, H., Meyer, H. E., Warscheid, B., Sa-Miranda, C., and Azevedo, J. E. (2008) J. Biol. Chem. 283(21), 14190-14197 Schliebs, W., Girzalsky, W., and Erdmann, R. (2010) Nat. Rev. Mol. Cell Biol. 11(12), 885-890 Kragt, A., Voorn-Brouwer, T. M., Van den Berg, M., and Distel, B. (2005) J. Biol. Chem. 280(8), 7867-7874 Platta, H. W., El Magraoui, F., Bäumer, B. E., Schlee, D., Girzalsky, W., and Erdmann, R. (2009) Mol. Cell. Biol. 29(20), 5505-5516 11 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 References 23. 24. 25. 26. 27. 28. 29. 30. 33. 34. 35. 36. 37. 38. 39. 40. 41. 42. 43. 44. 45. 46. 47. 48. 49. 12 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 31. 32. Williams, C., van den Berg, M., Geers, E., and Distel, B. (2008) Biochem. Biophys. Res. Commun. 374(4), 620-624 Komander, D., Clague, M. J., and Urbe, S. (2009) Nat. Rev. Mol. Cell. Biol. 10(8), 550-563 Wilkinson, K. D. (1997) Faseb J 11(14), 1245-1256 Rumpf, S., and Jentsch, S. (2006) Mol. Cell. 21(2), 261-269 Grou, C. P., Carvalho, A. F., Pinto, M. P., Huybrechts, S. J., Sa-Miranda, C., Fransen, M., and Azevedo, J. E. (2009) J. Biol. Chem. 284(16), 10504-10513 Güldener, U., Heck, S., Fiedler, T., Beinhauer, J., and Hegemann, J. H. (1996) Nucleic Acids Res. 24(13), 2519-2524 Girzalsky, W., Rehling , P., Stein, K., Kipper, J., Blank, L., Kunau, W.-H., and Erdmann, R. (1999) J. Cell Biol. 144(6), 1151-1162 Heinemeyer, W., Kleinschmidt, J. A., Saidowsky, J., Escher, C., and Wolf, D. H. (1991) The EMBO journal 10(3), 555-562 Vojtek, A. B., Hollenberg, S. M., and Cooper, J. A. (1993) Cell 74(1), 205-214 Erdmann, R., Wiebel, F. F., Flessau, A., Rytka, J., Beyer, A., Fröhlich, K. U., and Kunau, W.-H. (1991) Cell 64(3), 499-510 Liu, C., Apodaca, J., Davis, L. E., and Rao, H. (2007) BioTechniques 42(2), 158, 160, 162 Birschmann, I., Rosenkranz, K., Erdmann, R., and Kunau, W.-H. (2005) FEBS J. 272(1), 47-58 Melcher, K. (2000) Analytical biochemistry 277(1), 109-120 Niedenthal, R. K., Riles, L., Johnston, M., and Hegemann, J. H. (1996) Yeast 12(8), 773-786 Birschmann, I., Stroobants, A. K., Van Den Berg, M., Schäfer, A., Rosenkranz, K., Kunau, W. H., and Tabak, H. F. (2003) Mol. Biol. Cell. 14(6), 2226-2236 Stein, K., Schell-Steven, A., Erdmann, R., and Rottensteiner, H. (2002) Mol. Cell. Biol. 22(17), 6059-6069 Elgersma, Y., Kwast, L., van den Berg, M., Snyder, W. B., Distel, B., Subramani, S., and Tabak, H. F. (1997) EMBO J. 16(24), 7326-7341 Chevray, P. M., and Nathans, D. (1992) Proc. Natl. Acad. Sci. USA 89(13), 5789-5793 Guarente, L. (1983) Methods Enzymol. 101, 181-189 Erdmann, R., Veenhuis, M., Mertens, D., and Kunau, W.-H. (1989) Proc. Natl. Acad. Sci. USA 86(14), 5419-5423 Grunau, S., Lay, D., Mindthoff, S., Platta, H. W., Girzalsky, W., Just, W., and Erdmann, R. (2011) Biochem. J. 434(1), 161-170 Messick, T. E., Russell, N. S., Iwata, A. J., Sarachan, K. L., Shiekhattar, R., Shanks, J. R., Reyes-Turcu, F. E., Wilkinson, K. D., and Marmorstein, R. (2008) J. Biol. Chem. 283(16), 11038-11049 Neuhoff, V., Stamm, R., Pardowitz, I., Arold, N., Ehrhardt, W., and Taube, D. (1990) Electrophoresis 11(2), 101-117 Wiese, S., Gronemeyer, T., Ofman, R., Kunze, M., Grou, C. P., Almeida, J. A., Eisenacher, M., Stephan, C., Hayen, H., Schollenberger, L., Korosec, T., Waterham, H. R., Schliebs, W., Erdmann, R., Berger, J., Meyer, H. E., Just, W., Azevedo, J. E., Wanders, R. J., and Warscheid, B. (2007) Mol. Cell. Proteomics. 6(12), 2045-2057 Agne, B., Meindl, N. M., Niederhoff, K., Einwächter, H., Rehling, P., Sickmann, A., Meyer, H. E., Girzalsky, W., and Kunau, W. H. (2003) Mol. Cell 11(3), 635-646 Albertini, M., Rehling, P., Erdmann, R., Girzalsky, W., Kiel, J. A. K. W., Veenhuis, M., and Kunau, W.-H. (1997) Cell 89(1), 83-92 Rottensteiner, H., Stein, K., Sonnenhol, E., and Erdmann, R. (2003) Mol. Biol. Cell. 14(10), 4316-4328 50. 51. 52. 53. 54. 55. 56. 57. 59. 60. 61. 62. 63. 64. 65. 66. 67. 68. 69. 70. 71. 72. 73. 74. 75. 13 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 58. Ellison, M. J., and Hochstrasser, M. (1991) J. Biol. Chem. 266(31), 21150-21157 Zhu, Q., Hulen, D., Liu, T., and Clarke, M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94(14), 7308-7313 Vornlocher, H. P., Hanachi, P., Ribeiro, S., and Hershey, J. W. (1999) J. Biol. Chem. 274(24), 16802-16812 Amerik, A. Y., Li, S. J., and Hochstrasser, M. (2000) Biological chemistry 381(9-10), 981-992 Lin, C. H., MacGurn, J. A., Chu, T., Stefan, C. J., and Emr, S. D. (2008) Cell 135(4), 714-725 Entian, K. D., Vogel, R. F., Rose, M., Hofmann, L., and Mecke, D. (1988) FEBS Lett. 236(1), 195-200 Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425, 686-691 Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S. P. (2003) Nat. Biotechnol. 21(8), 921-926 Tamura, S., Yasutake, S., Matsumoto, N., and Fujiki, Y. (2006) J. Biol. Chem., in press Papa, F. R., and Hochstrasser, M. (1993) Nature 366(6453), 313-319 Hochstrasser, M. (1996) Annu. Rev. Genet. 30(405-439) Hershko, A., and Rose, I. A. (1987) Proceedings of the National Academy of Sciences of the United States of America 84(7), 1829-1833 van der Leij, I., Franse, M. M., Elgersma, Y., Distel, B., and Tabak, H. F. (1993) Proc. Natl. Acad. Sci. USA 90(24), 11782-11786 Purdue, P. E., and Lazarow, P. B. (2001) J. Biol. Chem. 276(50), 47684-47689 Amerik, A., Swaminathan, S., Krantz, B. A., Wilkinson, K. D., and Hochstrasser, M. (1997) The EMBO journal 16(16), 4826-4838 Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A. P., Punna, T., Peregrin-Alvarez, J. M., Shales, M., Zhang, X., Davey, M., Robinson, M. D., Paccanaro, A., Bray, J. E., Sheung, A., Beattie, B., Richards, D. P., Canadien, V., Lalev, A., Mena, F., Wong, P., Starostine, A., Canete, M. M., Vlasblom, J., Wu, S., Orsi, C., Collins, S. R., Chandran, S., Haw, R., Rilstone, J. J., Gandi, K., Thompson, N. J., Musso, G., St Onge, P., Ghanny, S., Lam, M. H., Butland, G., Altaf-Ul, A. M., Kanaya, S., Shilatifard, A., O'Shea, E., Weissman, J. S., Ingles, C. J., Hughes, T. R., Parkinson, J., Gerstein, M., Wodak, S. J., Emili, A., and Greenblatt, J. F. (2006) Nature 440(7084), 637-643 Jamieson, D. J. (1998) Yeast 14(16), 1511-1527 Izawa, S., Inoue, Y., and Kimura, A. (1995) FEBS Lett. 368(1), 73-76 Smith, J. J., Ramsey, S. A., Marelli, M., Marzolf, B., Hwang, D., Saleem, R. A., Rachubinski, R. A., and Aitchison, J. D. (2007) Mol. Syst. Biol. 3, 115 Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000) Molecular biology of the cell 11(12), 4241-4257 Swaminathan, S., Amerik, A. Y., and Hochstrasser, M. (1999) Mol. Biol. Cell 10(8), 2583-2594 Jentsch, S., and Rumpf, S. (2007) Trends Biochem. Sci. 32(1), 6-11 Zhang, J. W., Han, Y., and Lazarow, P. B. (1993b) J. Cell. Biol. 123(5), 1133-1147 Legakis, J. E., Koepke, J. I., Jedeszko, C., Barlaskar, F., Terlecky, L. J., Edwards, H. J., Walton, P. A., and Terlecky, S. R. (2002) Mol. Biol. Cell 13(12), 4243-4255 Terlecky, S. R., Koepke, J. I., and Walton, P. A. (2006) Biochimica et biophysica acta 1763(12), 1749-1754 Petriv, O. I., and Rachubinski, R. A. (2004) J Biol Chem 279(19), 19996-20001 76. 77. 78. 79. Gabaldon, T., Snel, B., van Zimmeren, F., Hemrika, W., Tabak, H., and Huynen, M. A. (2006) Biology direct 1, 8 Doss-Pepe, E. W., Stenroos, E. S., Johnson, W. G., and Madura, K. (2003) Mol. Cell. Biol. 23(18), 6469-6483 Wang, Q., Li, L., and Ye, Y. (2008) J. Biol. Chem. 283(12), 7445-7454 Ernst, R., Mueller, B., Ploegh, H. L., and Schlieker, C. (2009) Mol. Cell 36(1), 28-38 FIGURE LEGENDS Figure 1. The yeast AAA-complex possesses ubiquitin hydrolase activity. MonoUb-Pex5pcontaining wild-type membranes were incubated with the AAA-complex purified from a cytosolic fraction of pex5Δ cells using the TEV-ProtA tag. Where indicated, the AAA-complex was preincubated with NEM or with Ub-aldehyde to inhibit the observed ubiquitin hydrolase activity. Figure 3. Ubp15p is partially localized to peroxisomes. (A) A cell-free extract of oleate-induced wild-type cells expressing genomically tagged Ubp15p (Ubp15p-TEV-ProtA) was separated by density gradient centrifugation (2.25%-24% Optiprep, 18% sucrose). Fractions were subjected to measurements of the activity of catalase and cytochrom c oxidase as peroxisomal or mitochondrial marker, respectively (upper panel). Equal portions of fractions were probed by immunoblotting (lower panel) with antibodies against the protein A tag, Pex13p (peroxisomes); Porin (mitochondria) as well as Fbp1p (cytosol). (B) Wild-type cells expressing both the PTS2 marker protein PTS2-dsRed as well as GFP-Ubp15p, with and without overexpression of Pex15p, were grown on oleic acid plates for two days and examined by fluorescence microscopy. While only a small portion of GFP-Ubp15p is localized to peroxisomes in cells containing normal levels of Pex15p, a higher fraction of the fusion protein was recruited to peroxisomes upon overexpression of Pex15p, indicated by the co-localization of GFP-Ubp15p and the peroxisomal dsRed-marker. Figure 4. The first AAA-domain of Pex6p mediates the Ubp15p-interaction. The L40 reporter yeast cells were cotransformed with empty two-hybrid plasmids pPC86 and pPC97 (~) or plasmids expressing indicated proteins. Double-transformants were lysed and subjected to liquid ßgalactosidase assay. ß-galactosidase activities (expressed in arbitrary units) indicate binding and are represented as mean values of three independent experiments performed in duplicate. Error bars denote SEM (standard error of the mean). Abbreviations: Ub= ubiquitin; N= amino-terminal domain; D1= first AAA domain; D2= second AAA domain. Figure 5. Ubp15p is an ubiquitin hydrolase acting on poly- as well as monoubiquitinated Pex5p. (A) PolyUb-chains were incubated with recombinant wild-type Ubp15p or mutant Ubp15p(C214) harbouring a substitution of the supposedly active site cysteine. At indicated time-points reactions were stopped by adding SDS-sample buffer. Equal amounts of the samples were subjected to SDSPAGE. The presence of the indicated proteins was monitored by immunoblotting with antibodies against ubiquitin or Ubp15p as indicated. Membranes isolated from (B) NEM-treated wild-type cells which harbour monoubiquitinated Pex5p were incubated with recombinant Ubp15p followed by Pex5p-immunoisolation or (C) pex1Δpex6Δ cells which contain poly-ubiquitinated Pex5p were incubated with recombinant Ubp15p without further purification steps. The presence of either NEM or 14 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 Figure 2. Ubp15p forms a complex with the AAA-peroxins. Protein complexes were isolated by affinity chromatography from soluble fractions of (A) protease deficient yeast strain cl3-ABYS-86 with His6-Pex6p or (B) from UTL-7A strain with endogenously encoded Pex1p fused to TEV-ProtA tag. For the latter, the untransformed strain served as control for the specificity of the isolation. Isolated proteins were visualized by silver stain or colloidal Coomassie as indicated. (C) His6-GSTUbp15p was isolated form soluble fraction of cl3-ABYS-86 strain and analyzed by immunoblotting. Equal volumes of load and the 100 x-concentrated eluate fractions were probed with antibodies raised against indicated proteins. The detection of cytosolic fructose1,6-bisphosphatase (Fbp1p) served as control for unspecific binding. Ub-aldehyde inhibits hydrolase activity of Ubp15p and serves as control. Samples were subjected to SDS-PAGE and immunoblot analysis and with antibodies against ubiquitin or Pex5p-specific antibodies as indicated to monitor the presence of ubiquitinated Pex5p. Figure 7. ubp15Δ-cells exhibit a lower steady state concentration of Pex5p but higher rate of ubiquitinated Pex5p. (A) Whole cell lysates of oleic acid-induced wild-type as well as ubp15Δ cells were prepared and subjected to immunoblot analysis with antibodies specific for Pex5p and mitochondrial porin, which served as loading control (left). Signal intensity was estimated by densitometric analysis (right). (B) Indicated strains were grown for 10h under oleic-acid conditions and for additional 4 h under same conditions in the presence of MG132 to inhibit proteasomal degradation. Whole cell lysates were prepared and equal portion were subjected to immunoblot analyses with Pex5p antibodies (left). Signal intensity of modified Pex5p in ubp15pΔ cells and unmodified Pex5p in wild-type was quantified by densitometry (right). Figure 8. ubp15Δ-cells exhibit oxidative-stress related protein import deficiencies and defective growth on oleic acid (A) The PTS1 marker protein GFP-PTS1 was transformed into wild-type and indicated mutant strains. The transformed strains were grown in liquid oleic acid media in the absence or presence of 0.2 mM H2O2. All strains exhibit normal import of the marker protein GFP-PTS1 on oleic acid medium without oxidative stress. Upon supplementation of the oleic acid medium with H2O2, the marker protein was partially mislocalized to the cytosol in both doa4∆ and upb15Δ cells whereas wild-type and ubp14∆ remained unaffected. (B) Indicated strains were cultured in either oleic acid or glucose as sole carbon source in the absence (closed symbols) and presence (open symbols) of 0.2 mM H2O2. At different time points samples were taken and optical density was estimated at 600 nm. 15 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 Figure 6. ubp15Δ-cells contain functional but clustered peroxisomes (A) Indicated strains were spotted as a series of 10-fold dilutions on media containing oleic acid as sole carbon source and incubated for 5 days at 30°C. In contrast to pex5Δ, ubp15Δ grew at wild-type rate, suggesting that the cells are not affected in peroxisome function. (B) The PTS1 marker protein GFP-SKL and the PTS2 marker protein PTS2-dsRed were co-transformed in wild-type, pex5∆ and ubp15∆-cells. The transformed strains were grown on oleic acid plates for two days and examined by fluorescence microscopy. Mutant pex5∆ cells are capable to import PTS2-proteins properly (indicated by the punctuate pattern) but are impaired in PTS1-dependent matrix protein import and accordingly mislocalize the marker protein to the cytosol. Both wild-type and ubp15∆ exhibit a punctuate congruent staining for both peroxisomal markers, indicative for normal peroxisomal protein import. The peroxisomes of ubp15∆ cells form clusters. (C) Ultrastructural appearance of clustered peroxisomes in ubp15Δ cells. Wild-type and ubp15∆-mutant cells were grown on oleic acid medium and analyzed by electron microscopy. In wild-type cells, peroxisomes are separated and distributed within the cell, whereas the ubp15Δ mutant cells are characterized by peroxisome clusters. Peroxisomes are marked with an asterisk. Size bar: 2.5 µm. Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 16 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 17 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 18 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 19 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 20 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 21 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 22 Downloaded from www.jbc.org at MEDIZINISCHE EINRICHTUNGE, on June 10, 2011 23 EUKARYOTIC CELL, June 2011, p. 770–775 1535-9778/11/$12.00 doi:10.1128/EC.05038-11 Copyright © 2011, American Society for Microbiology. All Rights Reserved. Vol. 10, No. 6 The Putative Saccharomyces cerevisiae Hydrolase Ldh1p Is Localized to Lipid Droplets䌤 Sven Thoms,1† Mykhaylo O. Debelyy,1 Melanie Connerth,2 Günther Daum,2 and Ralf Erdmann1* Abteilung für Systembiochemie, Institut für Physiologische Chemie, Medizinische Fakultät der Ruhr-Universität Bochum, D-44780 Bochum, Germany,1 and Institute of Biochemistry, Graz University of Technology, A-8010 Graz, Austria2 Received 18 March 2011/Accepted 30 March 2011 Here, we report the identification of a novel hydrolase in Saccharomyces cerevisiae. Ldh1p (systematic name, Ybr204cp) comprises the typical GXSXG-type lipase motif of members of the ␣/-hydrolase family and shares some features with the peroxisomal lipase Lpx1p. Both proteins carry a putative peroxisomal targeting signal type1 (PTS1) and can be aligned with two regions of homology. While Lpx1p is known as a peroxisomal enzyme, subcellular localization studies revealed that Ldh1p is predominantly localized to lipid droplets, the storage compartment of nonpolar lipids. Ldh1p is not required for the function and biogenesis of peroxisomes, and targeting of Ldh1p to lipid droplets occurs independently of the PTS1 receptor Pex5p. prevents peroxisomal localization (40). A peroxisomal targeting signal type 2 (PTS2) is located within the first 20 amino acids of the N terminus of some peroxisomal proteins. Peroxisomal proteins with a PTS2 are recognized by the import receptor Pex7p (20, 21, 42). Here, we report the identification of a novel hydrolase in S. cerevisiae. The gene sequence of LDH1 predicts a GXSXGtype motif that is typical of ␣/-hydrolases and/or lipases (31). Bioinformatics analysis suggests that LDH1 (YBR204C) encodes a novel peroxisomal protein, due to its putative PTS1 (17). In the present study, however, we show that Ldh1p is not required for the function and biogenesis of peroxisomes and that Ldh1p primarily localizes to LDs, independently of the peroxisomal protein import machinery. Peroxisomes and lipid droplets (LDs) are ubiquitous eukaryotic organelles involved in lipid metabolism. LDs appear as oleosomes in plants, as adiposomes in mammals, or as lipid particles/bodies/droplets in yeasts and constitute a family of morphologically and biogenetically similar organelles (19). LDs are bound by a phospholipid monolayer and serve as the main storage sites for nonpolar lipids, mainly triacylglycerols (TAG) and cholesteryl ester (CE) (6, 7). LDs derive from the endoplasmic reticulum (ER), possibly by inclusion of nonpolar lipids between the two ER leaflets, eventually leading to the budding of nascent LDs (1, 6, 24, 27, 36). A large number of LD proteins have been identified by proteomic studies (12). In recent years, it has become evident that LDs, rather than being solely lipid storage sites, play a dynamic role in lipid biosynthesis, metabolism, degradation, and trafficking (6). Peroxisomes are particularly engaged in the -oxidation of long- and very long-chain fatty acids (16). Notably, in yeast, peroxisomes are the only site of fatty acid -oxidation (37). In mammals, peroxisomes are also involved in bile acid and plasmalogen synthesis, as well as amino acid metabolism (37, 38). Defective peroxisome biogenesis can lead to severe heritable diseases in humans (32). Such biogenesis defects are caused by mutations in PEX genes coding for proteins required for peroxisome biogenesis, collectively called peroxins (25, 34). The majority of peroxisomal matrix proteins are directed to peroxisomes by a peroxisomal targeting signal type1 (PTS1). The three amino acids SKL (serine-lysine-leucine) at the very C terminus of a protein represent the first PTS1 discovered. Generally, PTS1 comprises tripeptides with the consensus sequence [SAC] [KRH][LM]. The PTS1 is recognized in the cytosol by the cycling import receptor Pex5p (8). Masking of the PTS1 by the addition of protein tags interrupts PTS1-Pex5p association and MATERIALS AND METHODS Strains and plasmids. S. cerevisiae strains BY4742, BY4742 ⌬yor084w, BY4742 ⌬ybr204c, BY4742 ⌬pex5, and BY4742 ⌬pex1 were obtained from EUROSCARF (Frankfurt). BY4742 ERG6-RFP was obtained from W. K. Huh (San Francisco, CA). BY4742 ERG6-RFP ⌬ybr204c was constructed by gene replacement using kanMX6 from pUG6 and primers 5⬘-CTAGAAGAGATTG TTCAAAATGCAGAAAATGCAGCTGATTTGGTCGTACGCTGCAGGTC GAC-3⬘ and 5⬘-GCACGAAAATCTAGTTACGCAATGTGAAATCTAGAAA ACCTTCTAATCGATGAATTCGAGCTCG-3⬘. BY4742 ⌬pex5⌬ldh1 and BY4742 ⌬pex1⌬ldh1 were constructed from BY4742 ⌬pex5 and BY4742 ⌬pex1 by gene replacement using a pUG6 vector and primers 5⬘-GCTAGAAGAGATTG TTCAAAATGCAGAAAATGCAGCTGATTTGGTCGTACGCTGCAGGTC GAC-3⬘ and 5⬘-GCACGAAAATCTAGTTACGCAATGTGAAATCTAGAAA ACCTTCTAATCGATGAATTCGAGCTCG-3⬘ after removal of loxPkanMX6-loxP marker cassettes (13, 14). The yeast media have been described previously (9, 10). For construction of pUG35-LDH1 (Ldh1p-GFP), PCR-amplified YBR204c (primers RE2444 [5⬘-GCGCGGATCCATGAATATGGCAG AACGTGCA-3⬘] and RE2445 [5⬘-GCGCAAGCTTCAATTTGGAATTATCA ATCAC-3⬘]) was introduced into BamH I and HindIII sites of pUG35. For construction of pUG36-LDH1 (GFP-Ldh1p), PCR-amplified YBR204C (primers RE2444 [5⬘-GCGCGGATCCATGAATATGGCAGAACGTGCA-3⬘] and RE2446 [5⬘-GCGCAAGCTTCTACAATTTGGAATTATCAATCAC-3⬘]) was introduced into BamHI and HindIII sites of pUG36. All constructs were confirmed by DNA sequencing. The GFP-SKL plasmid has been described previously (29). Nile Red and Oil Red O staining. For Nile Red staining (39), yeast cells in stationary phase were washed and resuspended in phosphate-buffered saline (PBS) (150 mM NaCl, 1.7 mM KH2PO4, 5.2 mM Na2HPO4). The cells were stained with Nile Red solution (0.0005% in PBS, diluted from a 0.01% stock solution in acetone) for 15 min at room temperature in the dark. The cells were * Corresponding author. Mailing address: Institut für Physiologische Chemie, Ruhr-Universität Bochum, Universitätsstr. 150, D-44780 Bochum, Germany. Phone: 49 234 322 4943. Fax: 49 234 321 4266. E-mail: [email protected]. † Present address: Universitätsmedizin Göttingen, Abteilung für Pädiatrie und pädiatrische Neurologie, Georg-August-Universität Göttingen, D-37099 Göttingen, Germany. 䌤 Published ahead of print on 8 April 2011. 770 VOL. 10, 2011 PUTATIVE YEAST HYDROLASE Ldh1p IS LOCALIZED TO LIPID DROPLETS 771 FIG. 1. Ldh1p and Lpx1p from S. cerevisiae are similar proteins with a hydrolase/lipase motif. (A) Similarities between Lpx1p (predicted mass, 43.7 kDa; 387 amino acids; theoretical pI, 8.16) and Ldh1p (predicted mass, 43.3 kDa; 375 amino acids; theoretical pI, 6.36) are indicated: two regions of homology, the first of which contains the GHSMG hydrolase/lipase motif of the GXSXG consensus. Both proteins carry a (putative) PTS1, QKL, or SKL. (B) Alignment of the two regions of homology of Lpx1p and Ldh1p exhibiting 28% (region A) and 27% (region B) amino acid identities. Asterisk, histidine of the probable catalytic triad; arrowhead, aspartate of the probable catalytic triad in Ldh1p. The GXSXG hydrolase/lipase motif is underlined; similar amino acids are indicated by a plus symbol. (C) Hydropathy plots of Ldh1p. The Kyte-Doolittle plot was calculated with a window size of 11. Values greater than 1.8 indicate very hydrophobic regions. (D) C terminus of Ldh1p. The amino acids in positions ⫺2 and ⫺5 are likely to interfere with peroxisomal targeting. then washed six times with PBS to remove surplus dye. For Oil Red O staining (26, 39), yeast cells in stationary phase were washed twice, fixed by 4% formaldehyde in PBS for 20 min, and washed twice again. The cells were then stained with Oil Red O (0.2% in a water-isopopanol [1:1] mixture) for 15 min at room temperature in the dark and washed six times before microscopic analysis. Image acquisition. Samples were fixed with 0.5% (wt/vol) agarose on microscope slides. Fluorescence microscopic images were recorded on an AxioPlan 2 microscope (Zeiss) equipped with a ␣Plan-FLUAR 100⫻/1.45 oil objective and an AxioCam MRm camera (Zeiss) at room temperature. If necessary, contrast was linearly adjusted using the image acquisition software AxioVision 4.8 (Zeiss). Subcellular fractionation and organelle isolation. Subcellular fractionation and gradient centrifugation for the analysis of peroxisomes and mitochondria of ⌬ldh1 were carried out as described previously (29, 33). Cell fractionation and LD isolation for the subcellular localization of Ldh1p have been described previously (5, 11, 28). RESULTS Ldh1p and Lpx1p: two similar hydrolases. Ldh1p shares some features with the peroxisomal lipase Lpx1p (33) (Fig. 1). Both proteins have almost the same predicted molecular mass, namely, 43 kDa for Ldh1p and 44 kDa for Lpx1p. Both proteins carry a putative PTS1, the prototypical SKL in Ldh1p, and glutamine-lysine-leucine (QKL) in Lpx1p (Fig. 1A). Furthermore, both proteins can be aligned with two regions of homology (Fig. 1A and B), with one in the central domain, comprising the lipase motif GHSMG (4, 35), indicative of members of the ␣/-hydrolase family. In the case of Ldh1p, the amino acids adjacent to the active-site serine are identical in the two proteins, namely, histidine (H) and methionine (M). Hydropathy plots indicated a pronounced hydrophobic region in the centers of both proteins. Amino acids 130 to 154 of Ldh1p comprise a hydrophobic core region, 138VVELIFVLV 146, and amino acids 154 to 177 of Lpx1p comprise the core region, 164LLILIEPVVI173 (Fig. 1C). Absence of a synthetic phenotype of ⌬ldh1 and ⌬lpx1 in peroxisome biogenesis. Ldh1p carries the prototypical yet putative PTS1 and has been speculated to be a peroxisomal matrix protein (17). Therefore, we first tested the effect of an LDH1 deletion on peroxisome biogenesis. Postnuclear supernatants (PNS) were prepared from wild-type and ⌬ldh1 strains and analyzed by density gradient centrifugation. The gradient fractions were assayed for peroxisomal catalase and mitochondrial cytochrome c oxidase activity (Fig. 2A). The distribution of neither of these proteins indicated a significant change in the abundance or density of peroxisomes or mitochondria, suggesting that peroxisomal and mitochondrial biogenesis remain functional after deletion of LDH1. As a defect in peroxisome biogenesis would affect peroxisome presence or density, we conclude that Ldh1p is not a peroxin. Altogether, the avail- 772 EUKARYOT. CELL THOMS ET AL. FIG. 2. Ldh1p is dispensable for peroxisome biogenesis and function. (A) Postnuclear supernatants prepared from oleate-induced wildtype and ⌬ldh1 strains were fractionated by density gradient centrifugation, and each fraction was analyzed for catalase (peroxisome) and cytochrome c oxidase (mitochondria) activities. The absence of Ldh1p has no influence on the apparent densities of peroxisomes and mitochondria. (B) Growth on oleate is not affected by deletion of the lipase gene LDH1 or LPX1 or both. Single or double deletions of LDH1 and LPX1 were spotted on oleate and ethanol plates with equal cell numbers in a series of 10-fold dilutions and grown for 3 days at 30°C. able evidence suggested that Lpx1p and Ldh1p might be proteins exerting similar or redundant functions. Most mutants whose peroxisome biogenesis or functions are affected are characterized by a growth defect on oleic acid (9). We therefore tested the single and double knockouts of LPX1 and LDH1 for growth on oleate as the only carbon source (Fig. 2B). Neither of these knockouts had its growth on oleic acid affected, suggesting that Lpx1p and Ldh1p do not form a redundant pair in peroxisome function. Ldh1p localizes to the lipid droplet membrane. Next, we investigated the subcellular distribution of Ldh1p. Ldh1p was expressed from a plasmid as N-terminally or C-terminally tagged green fluorescent protein (GFP) fusion proteins that localized to a particular organelle about 1 to 2 m in diameter with several copies in a cell (Fig. 3A). Ldh1p specifically local- ized to the surface membranes of these organelles. We reasoned that the organelles were fragmented vacuoles, endosomes, or LDs. Thus, we coexpressed marker proteins for the organelles together with the Ldh1p fusion proteins and found that Ldh1p perfectly colocalized with Erg6p, the ␦(24)-sterol methyl transferase (Fig. 3A, top), which is a major and prominent LD protein (18). Both proteins localize to the surface membrane of LDs. Ldh1p colocalized with Erg6p when GFP was localized at the N terminus or the C terminus of the protein (Fig. 3A). Localization of Ldh1p in LDs was also confirmed by Oil Red O staining (Fig. 3B). Ldh1p contains a perfect consensus for a PTS1 at its extreme C terminus. The fact that some LD proteins contain a C-terminal localization signal (22) and the possibility of a common origin of peroxisomes and LD encouraged us to test whether the PTS1 of Ldh1p is required for LD targeting. We found that neither masking of the SKL by expression of the GFP at the C terminus of Ldh1p nor deletion of the PTS1 receptor protein Pex5p interfered with targeting of Ldh1p (Fig. 3C). Thus, the PTS1-like C terminus of Ldh1p does not function as a classical peroxisomal targeting signal, nor does it interfere with targeting of the polypeptide to LD. To verify the localization of Ldh1p, we performed cell fractionation analysis with a yeast strain that expressed plasmidencoded Ldh1p-GFP. LDs were isolated by flotation on a density gradient (5, 28). Subcellular fractions of the gradient were analyzed by immunoblotting with polyclonal antibodies against GFP and organelle-specific marker enzymes (Fig. 4). These data revealed that Ldh1p-GFP was highly enriched in LD, as represented by the LD marker proteins Erg1p (squalene epoxidase) and Erg6p, but Ldh1p-GFP also cofractionated to some extent with the peroxisomal marker protein Fox1p (fatty-acyl coenzyme A oxidase) and the mitochondrial marker protein Por1p (mitochondrial porin) (Fig. 4). It has been shown that some LD proteins are not exclusively found in this compartment but also localize to the ER; in contrast, Ldh1p appears to localize to LD and, possibly to a lesser extent, to mitochondria and peroxisomes. The biogenesis of peroxisomes and lipid droplets does not require LDH1. To test whether deletion of LDH1 influences the intracellular distribution or morphology of peroxisomes, we analyzed wild-type and ⌬ldh1 strains expressing the peroxisomal marker protein GFP-SKL by fluorescence microscopy. Microscopic inspection of the LD was performed by Oil Red O staining (Fig. 5). These results showed that the morphological appearance of peroxisomes, as well as the frequently observed proximity to LD, was not affected by deletion of LDH1. Having shown that Ldh1p is targeted to LD independently of the soluble PTS1 receptor, we investigated whether Ldh1p is required for the biogenesis of LDs. After introducing a ⌬ldh1 knockout into the genomically tagged ERG6-red fluorescent protein (RFP) marker strain for LD, we found that LD could still be formed in the absence of Ldh1p (Fig. 6A). We confirmed these findings by LD staining with Nile Red (Fig. 6B) and Oil Red O (Fig. 6C). Taking these data together, it appears that Ldh1p is not required for the formation of LD. DISCUSSION Ldh1p is a lipid droplet hydrolase with an SKL terminus. Ldh1p contains the consensus sequence for a classical peroxi- VOL. 10, 2011 PUTATIVE YEAST HYDROLASE Ldh1p IS LOCALIZED TO LIPID DROPLETS 773 FIG. 3. Ldh1p primarily localizes to lipid droplets, and its localization is independent of the peroxisomal import receptor Pex5p. (A) Ldh1p colocalizes with the LD marker protein Erg6p [␦(24)-sterol methyl transferase]. GFP-Ldh1p and Ldh1p-GFP were coexpressed in a yeast strain with genomically tagged Erg6p-RFP. Bar, 1 m. (B) Ldh1p colocalizes with the LD marker dye Oil Red O. GFP-Ldh1p and Ldh1p-GFP were coexpressed in a wild-type yeast strain. Bar, 1 m. (C) Ldh1p localization is independent of the peroxisomal PTS1 pathway. GFP was fused to either the C terminus (top images) or the N terminus (bottom images) of Ldh1p. Also, in a ⌬pex5 deletion mutant, Ldh1p localization to LD was not compromised (right). In both cases Ldh1p colocalizes with the LD marker dye Oil Red O. somal targeting signal, but the protein is primarily targeted to LD and not to peroxisomes. Peroxisomal exclusion of Ldh1p is likely due to the upstream sequences with charged amino acids in positions ⫺2 and ⫺5 (Fig. 1D). These positions are adverse to Pex5p binding and peroxisomal localization, for which polar/ hydrophilic or positively charged amino acids in position ⫺2 FIG. 4. Subcellular localization of Ldh1p. (A) Organelles from the wild-type strain carrying Ldh1p-GFP were isolated from cells grown to stationary phase in oleic acid-containing medium. Proteins from the subcellular fractions were precipitated, and the same amounts were separated by SDS-PAGE and analyzed by Western blotting using primary antibodies against marker enzymes, as indicated. The same amounts of proteins were loaded; therefore, the intensity of the GFP band does not represent the relative distribution of Ldh1p between LDs, mitochondria, and peroxisomes. The presence of organelles was detected with primary antibody against marker enzymes, as indicated. Erg1p, squalene epoxidase; Erg6p, ␦(24)-sterol methyl transferase (lipid droplets); Fox1p, fatty-acyl coenzyme A oxidase (peroxisomes); Por1p, porin (mitochondria); Wbp1p (endoplasmic reticulum); H, homogenate; C, cytosol; 40g, 40,000 ⫻ g microsomes (endoplasmic reticulum); 100g, 100,000 ⫻ g microsomes (endoplasmic reticulum); Mt, mitochondria; Px, peroxisomes. are preferred. In our case, the negatively charged amino acid is not even counteracted by neighboring amino acids, giving the likely explanation for dominating peroxisomal exclusion. The classical PTS1, SKL, is not completely sufficient to target protein to peroxisomes if the upstream sequences are not supportive. We show that the majority of Ldh1p is an LD protein that is targeted independently of the PTS1-binding Pex5p. This view is confirmed by applying a PTS1 prediction algorithm FIG. 5. The association of lipid droplets and peroxisomes is not affected by deletion of LDH1. Shown is fluorescence microscopy of wild-type yeast and the ⌬ldh1 strain transformed with pGFP-SKL. LDs were stained with Oil Red O (ORO). BF, bright field. Bar, 1 m. 774 EUKARYOT. CELL THOMS ET AL. Extended localization studies of Ldh1p-GFP showed that at least a portion of the polypeptide is targeted to peroxisomes and mitochondria. While this triple localization may reflect the true cellular scenario, we also have to take into account that partial targeting of Ldh1p to peroxisomes and mitochondria may be due to the overexpression of Ldh1p-GFP. We were able to show that Ldh1p and the lipase Lpx1p are not redundant, provided that other enzymes, probably with somewhat lower homology, cannot compensate for a defect in the two enzymes. Both peroxisomes and LD function in concert in lipid metabolism. LDs require the action of triacylglycerol lipases to metabolize nonpolar lipids, while peroxisomes represent the sole cellular site for fatty acid oxidation. It is thus possible that the peroxisomal Lpx1p and the LD Ldh1p play a physiological role in lipid metabolism by mobilizing fatty acids and channeling them to their site of degradation. LDs, as fatty acid depot organelles, can be the storage sites for nonpolar lipids that are further metabolized in peroxisomes. For this reason, and not surprisingly, LDs have been found in proximity to peroxisomes in different organisms (2, 15, 30). It was also shown that S. cerevisiae peroxisomes attach to LDs or even project into LDs, which was interpreted as an intimate interaction between the two compartments (3). Our work on Ldh1p and Lpx1p shows that, beyond a metabolic collaboration, peroxisomes and LDs may be equipped with similar hydrolases. ACKNOWLEDGMENTS We thank Elisabeth Becker, Monika Bürger, and Uta Ricken for technical assistance; Robert Rucktäschel for scientific input; and Wolfgang Girzalsky for reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB642, ER178/4-1). REFERENCES FIG. 6. Lipid droplet biogenesis is independent of Ldh1p. (A) Comparison of Erg6p-RFP localization in a wild-type strain and a ⌬ldh1 deletion strain. Bar, 2 m. (B) Localization and morphology of Nile Red-stained LDs from the wild type and a ⌬ldh1 deletion strain. Bar, 1 m. (C) LD morphology in the wild type and ⌬lpx1, ⌬ldh1, and ⌬ldh1⌬lpx1 deletion strains. LDs were stained with Oil Red O. BF, bright field. Bar, 1 m. (http://mendel.imp.ac.at/pts1/) (23) that does not predict peroxisomal localization for Ldh1p. LD localization signals are only poorly characterized. It has been suggested that LD localization signals are constituted of hydrophobic residues at the C terminus of a protein (22, 41). A Kyte-Doolittle plot of Ldh1p indicated a region with particularly high hydrophobicity from amino acids 130 to 154 (Fig. 1C). This stretch might be required to target and/or to attach Ldh1p to LDs. Indeed, our data show that LD targeting is not abrogated when GFP is added to the C terminus or the N terminus of Ldh1p. Thus, targeting information within central parts of Ldh1p, rather than at its termini, is sufficient for the LD localization. Interestingly, the Lpx1p stretch of high hydrophobicity is in a similar location in the primary sequence, namely, at amino acids 154 to 177. The hydrophobic stretches in Ldh1p are likely not classical transmembrane domains (TMD), because LDs are bound by a single monolayer membrane of phospholipids. 1. Athenstaedt, K., and G. Daum. 2006. The life cycle of neutral lipids: synthesis, storage and degradation. Cell. Mol. Life Sci. 63:1355–1369. 2. Bascom, R. A., H. Chan, and R. A. Rachubinski. 2003. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants. Mol. Biol. Cell 14:939–957. 3. Binns, D., et al. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173:719–731. 4. Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528–530. 5. Connerth, M., K. Grillitsch, H. Kofeler, and G. Daum. 2009. Analysis of lipid particles from yeast. Methods Mol. Biol. 579:359–374. 6. Czabany, T., K. Athenstaedt, and G. Daum. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771:299–309. 7. Czabany, T., et al. 2008. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283:17065– 17074. 8. Dodt, G., et al. 1995. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9:115–125. 9. Erdmann, R., M. Veenhuis, D. Mertens, and W. H. Kunau. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86:5419–5423. 10. Erdmann, R., et al. 1991. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64:499–510. 11. Fowler, S. D., and P. Greenspan. 1985. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J. Histochem. Cytochem. 33:833–836. 12. Goodman, J. M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50:2148–2156. 13. Gueldener, U., J. Heinisch, G. J. Koehler, D. Voss, and J. H. Hegemann. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. 14. Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524. VOL. 10, 2011 PUTATIVE YEAST HYDROLASE Ldh1p IS LOCALIZED TO LIPID DROPLETS 15. Hayashi, Y., M. Hayashi, H. Hayashi, I. Hara-Nishimura, and M. Nishimura. 2001. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218:83–94. 16. Kunau, W. H., et al. 1988. Comparative enzymology of beta-oxidation. Biochem. Soc. Trans. 16:418–420. 17. Lazarow, P. B., and W. H. Kunau. 1997. Peroxisomes, p. 547–605. In E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 18. Leber, R., E. Zinser, G. Zellnig, F. Paltauf, and G. Daum. 1994. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast 10: 1421–1428. 19. Martin, S., and R. G. Parton. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378. 20. Marzioch, M., R. Erdmann, M. Veenhuis, and W. H. Kunau. 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13:4908–4918. 21. Miyata, N., K. Hosoi, S. Mukai, and Y. Fujiki. 2009. In vitro import of peroxisome-targeting signal type 2 (PTS2) receptor Pex7p into peroxisomes. Biochim. Biophys. Acta 1793:860–870. 22. Müllner, H., D. Zweytick, R. Leber, F. Turnowsky, and G. Daum. 2004. Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1663:9–13. 23. Neuberger, G., S. Maurer-Stroh, B. Eisenhaber, A. Hartig, and F. Eisenhaber. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328:581–592. 24. Ohsaki, Y., et al. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta 1791:399–407. 25. Platta, H. W., and R. Erdmann. 2007. The peroxisomal protein import machinery. FEBS Lett. 581:2811–2819. 26. Puri, V., et al. 2007. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J. Biol. Chem. 282:34213–34218. 27. Rajakumari, S., K. Grillitsch, and G. Daum. 2008. Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47:157–171. 28. Rosenberger, S., M. Connerth, G. Zellnig, and G. Daum. 2009. Phosphatidylethanolamine synthesized by three different pathways is supplied to peroxisomes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1791:379–387. 775 29. Schäfer, A., D. Kerssen, M. Veenhuis, W. H. Kunau, and W. Schliebs. 2004. Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24:8895–8906. 30. Schrader, M. 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49:1421–1429. 31. Schrag, J. D., and M. Cygler. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 284:85–107. 32. Steinberg, S. J., et al. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta 1763:1733–1748. 33. Thoms, S., M. O. Debelyy, K. Nau, H. E. Meyer, and R. Erdmann. 2008. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 275:504–514. 34. Thoms, S., and R. Erdmann. 2005. Import of proteins into peroxisomes, p. 125–134. In J. Eichler (ed.), Protein movement across membranes. Landes Bioscience, Georgetown, TX. 35. Voss, H., et al. 1997. DNA sequencing and analysis of 130 kb from yeast chromosome XV. Yeast 13:655–672. 36. Walther, T. C., and R. V. Farese, Jr. 2009. The life of lipid droplets. Biochim. Biophys. Acta 1791:459–466. 37. Wanders, R. J., and H. R. Waterham. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332. 38. Wanders, R. J., and H. R. Waterham. 2006. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta 1763:1707–1720. 39. Wolinski, H., and S. D. Kohlwein. 2008. Microscopic analysis of lipid droplet metabolism and dynamics in yeast. Methods Mol. Biol. 457:151–163. 40. Yang, X., P. E. Purdue, and P. B. Lazarow. 2001. Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 80:126–138. 41. Zehmer, J. K., R. Bartz, P. Liu, and R. G. Anderson. 2008. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J. Cell Sci. 121:1852–1860. 42. Zhang, J. W., and P. B. Lazarow. 1996. Peb1p (Pas7p) is an intraperoxisomal receptor for the NH2-terminal, type 2, peroxisomal targeting sequence of thiolase: Peb1p itself is targeted to peroxisomes by an NH2-terminal peptide. J. Cell Biol. 132:325–334. EUKARYOTIC CELL, June 2011, p. 776–781 1535-9778/11/$12.00 doi:10.1128/EC.05040-11 Copyright © 2011, American Society for Microbiology. All Rights Reserved. Vol. 10, No. 6 Involvement of the Saccharomyces cerevisiae Hydrolase Ldh1p in Lipid Homeostasis䌤 Mykhaylo O. Debelyy,1 Sven Thoms,1† Melanie Connerth,2 Günther Daum,2 and Ralf Erdmann1* Abteilung für Systembiochemie, Institut für Physiologische Chemie, Medizinische Fakultät der Ruhr-Universität Bochum, D-44780 Bochum, Germany,1 and Institute of Biochemistry, Graz University of Technology, A-8010 Graz, Austria2 Received 18 March 2011/Accepted 30 March 2011 Here, we report the functional characterization of the newly identified lipid droplet hydrolase Ldh1p. Recombinant Ldh1p exhibits esterase and triacylglycerol lipase activities. Mutation of the serine in the hydrolase/lipase motif GXSXG completely abolished esterase activity. Ldh1p is required for the maintenance of a steady-state level of the nonpolar and polar lipids of lipid droplets. A characteristic feature of the Saccharomyces cerevisiae ⌬ldh1 strain is the appearance of giant lipid droplets and an excessive accumulation of nonpolar lipids and phospholipids upon growth on medium containing oleic acid as a sole carbon source. Ldh1p is thought to play a role in maintaining the lipid homeostasis in yeast by regulating both phospholipid and nonpolar lipid levels. eration of energy by -oxidation or for the synthesis of membrane lipids and signaling molecules (9). It has been shown that nearly all cell types have the ability to generate LDs in response to elevated fatty acid levels and to subsequently metabolize and disperse these LDs when conditions are reversed (26), thereby providing an emergency energy pool for cell survival (3). Due to their unique architecture, LDs can protect cells from the effects of potentially toxic lipid species, such as unesterified lipids (23, 24) or toxic free fatty acids (3), by depositing them inside the LD’s core. In addition to this lipid-scavenging function, LDs can transiently store certain proteins, which may be released or degraded at later time points (9, 13, 14, 36). Here, we report the functional characterization of the newly identified LD hydrolase Ldh1p (34a). We demonstrate that recombinant Ldh1p exerts esterase and triacylglycerol lipase activities. The enzyme activity was abolished upon mutation of the conserved GXSXG-type lipase motif of the protein. The Saccharomyces cerevisiae ⌬ldh1 strain is characterized by the appearance of giant LDs and the accumulation of nonpolar lipids and phospholipids in LDs, indicative of a role of Ldh1p in maintaining lipid homeostasis. Lipid droplets (LDs) are remarkable dynamic subcellular organelles of globular shape with a size range from 20 to 100 m, depending on the cell type (9, 12, 15, 31). LDs are depots of neutral lipids with a complex biology that exist in virtually any kind of cell, ranging from bacteria to yeasts, plants, and higher mammals (3, 13, 15). In many cells, LDs occupy a considerable portion of the cell volume and weight (35). As the major intracellular storage organelles, LDs were first described in the works of R. Altmann and E. B. Wilson in the 19th century (1, 37). In contrast to the vesicular organelles, which have the aqueous content enclosed by a phospholipid bilayer membrane (12, 13), mature LDs have a unique physical structure: they have a neutral lipid core consisting of triacylglycerols (TG) and sterol esters (SE) surrounded by a phospholipid monolayer (3, 24, 38) that contains numerous peripheral or embedded proteins (26, 33). TG as well as SE play crucial roles for the cell: TG is the main energy store, and both TG and SE are depots of membrane lipid components (35). LDs can tightly regulate the level of intracellular free cholesterol by hydrolyzing sterol ester (26). The LD core also contains other endogenous neutral lipids, like monoacylglycerol, diacylglycerol, free cholesterol, and retinol ester, and xenobiotic hydrophobic compounds, such as polycyclic aromatic hydrocarbons (15, 17, 29, 32, 33). A number of proteins are specifically targeted to the LD surface (18), where they can regulate LD dynamics and the turnover of stored lipids (24). Lipid-metabolizing enzymes, including hydrolases and lipases, are the major class of LD enzymes (9). LDs play crucial roles in cellular energy homeostasis and lipid metabolism (35). LDs can provide a rapidly mobilized lipid source for many important biological processes. Neutral lipids may be mobilized for the gen- MATERIALS AND METHODS Strains and plasmids. S. cerevisiae strains BY4742, BY4742 ⌬ybr204c, BY4742 ⌬yor084w, BY4742 ⌬ybr204c ⌬yor084w, BY4742 ERG6-RFP, and BY4742 ERG6-RFP ⌬ybr204c are described in reference 34a. DNA plasmids pUG35LDH1 (Ldh1p-GFP) and pUG36-LDH1 (GFP-Ldh1p) are described in reference 34a. Yeast media have been described previously (10, 11). pUG35-LDH1-M1 [Ldh1p-(S177A)-GFP] and pUG36-LDH1-M1 [GFP-Ldh1p-(S177A)] were cloned from pUG35-LDH1 and pUG36-LDH1 using a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) (primers RE2400 [5⬘-ATAGTGCTTGTA GGGCATGCTATGGGTTGTTTTCTGGCA-3⬘] and RE2401 [5⬘-TGCCAGA AAACAACCCATAGCATGCCCTACAAGCACTAT-3⬘]). pET21d-LDH1 was constructed by introducing PCR-amplified YBR204c (primers OST248 [5⬘-GC GAATTCCATATGAATATGGCAGAACGTGCAG-3⬘] and OST217 [5⬘-GCT GCGGCCGCCAATTTGGAATTATCAATCACC-3⬘]) into NdeI and NotI sites of pET21b (EMD Chemicals). pET21d-LDH1-M1 [Ldh1p-(S177A)-His6] was cloned from pET21d-LDH1 using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) (primers RE2400 [5⬘-ATAGTGCTTGTAGGG CATGCTATGGGTTGTTTTCTGGCA-3⬘] and RE2401 [5⬘-TGCCAGAAAA * Corresponding author. Mailing address: Institut für Physiologische Chemie, Ruhr-Universität Bochum, Universitätsstraße 150, D-44780 Bochum, Germany. Phone: 49 234 322 4943. Fax: 49 234 321 4266. E-mail: [email protected]. † Present address: Universitätsmedizin Göttingen, Abteilung für Pädiatrie und pädiatrische Neurologie, Georg-August-Universität Göttingen, D-37099 Göttingen, Germany. 䌤 Published ahead of print on 8 April 2011. 776 VOL. 10, 2011 INVOLVEMENT OF YEAST HYDROLASE Ldh1p IN LIPID HOMEOSTASIS CAACCCATAGCATGCCCTACAAGCACTAT-3⬘]). All constructs were confirmed by DNA sequencing. Protein expression. Ldh1p was expressed from plasmid pET21b-LDH1 in Escherichia coli BL21(DE3). Cells were harvested by centrifugation and diluted in buffer A (1⫻ phosphate-buffered saline [PBS], 300 mM sodium chloride, 1 mM dithiothreitol, 40 mM imidazole) containing a protease inhibitor mixture (8 M antipain-dihydrochloride, 0.3 M aprotinin, 1 M bestatin, 10 M chymostatin, 5 M leupeptin, 1.5 M pepstatin), together with 50 g/ml lysozyme, 22.5 g/ml DNase I, and 40 mM imidazole. The cells were sonicated using a 250D Branson (Danbury, CT) Digital Sonifier. After removal of cell debris by centrifugation, the supernatant was clarified by 0.22-m filtration and loaded on HisTrap columns (GE Healthcare Life Sciences) equilibrated with buffer A. The column was washed in buffer A, and recombinant Ldh1p was eluted by a continuous 40 to 500 mM imidazole gradient. Peak fractions were identified by SDS-PAGE and pooled, and the isolated protein was concentrated with VivaSpin concentrators (30-kDa cutoff; Sartorius). The concentrated Ldh1p was subjected to size exclusion chromatography on an ÄKTA Purifier FPLC System with Superdex 200 (GE Healthcare Life Sciences). Peak fractions of Ldh1p were identified by SDS-PAGE and pooled, and the isolated protein was concentrated with VivaSpin (30-kDa cutoff; Sartorius). Enzyme assays. Esterase activity was determined with p-nitrophenyl butyrate (PNB) (Sigma) in PBS (pH 7.4) in a total volume of 200 l at 37°C. Free p-nitrophenol was determined at 410 nm in 96-well plates. Michaelis-Menten kinetics was analyzed using GraphPad Prism 5 (GraphPad Software). Triacylglycerol lipase (TGL) activity was determined using 1,2-dioleoyl-3-pyrenedecanol-rac-glycerol (DPG) (Marker Gene) in 0.1 M glycine, 19 mM sodium deoxycholate, pH 9.5, in a total volume of 200 l at 37°C. Hydrolysis of DPG was followed in 96-well plates at 460 nm with 360-nm excitation in a Sirius HT fluorescence plate reader (MWG Biotech). The TGL activity of Ldh1p toward DPG was compared with the TGL activity of Candida rugosa triacylglycerol lipase (Lipase AT30 Amano; 1,440 units/mg; Sigma) as a control. We also adapted a specific and sensitive TGL assay originally developed for the measurement of bacterial TGLs (22). The TGL activity of Ldh1p on rhodamine B agar plates was determined by using agar plates containing trioleoylglycerol and rhodamine B. The agar (1% [wt/vol]) was dissolved in PBS, adjusted to pH 7.4, autoclaved, and cooled to 60°C. Then, trioleoylglycerol (2.5% [wt/vol]) and rhodamine B (0.001% [wt/vol]) were added to the agar medium with vigorous stirring for 1 min. The medium was kept for 10 min at 60°C to reduce foaming, and 20 ml of medium was poured into plastic petri dishes. To detect triacylglycerol lipase activity, holes with a diameter of 6 mm were punched into the agar and filled with 200 l protein solution. Ldh1p and C. rugosa lipase (CRL) were diluted in PBS (pH 7.4). The plates were incubated for 48 h at 30°C. After 48 h, the plates began to show an orange fluorescence visible under UV light (350 nm). Lipid extraction and TLC. The lipids were extracted by the method of Bligh and Dyer (4). The organic layer was washed three times with 1 M KCl, and the solvent was removed by evaporation in a vacuum. The lipids were dissolved in a small volume of chloroform and separated on thin-layer chromatography (TLC) plates (TLC Silica gel 60 F254; 20 by 20 cm; Merck) using chloroformmethanol-water (65:25:4 [vol/vol/vol]) as the developing solvent. Lipid classes were visualized with iodine vapor and identified according to TLC standard 18-5A (Nu-Chek Prep, Elysian, MN). Electron microscopy. The ultrastructure of yeast cells was studied with oleateinduced cells that had been fixed with 1.5% KMnO4 and processed as described previously (10). Miscellaneous. Oil Red O staining, image acquisition, and the isolation of LDs are described in reference 34a. LD purification for lipid extraction was performed as described previously (8, 27). The weight of LDs was estimated gravimetrically in 1.5-ml reaction tubes (Eppendorf). 777 FIG. 1. Protein expression, purification, and enzymatic activity of Ldh1p. (A) Ldh1p was expressed as a fusion protein with a hexahistidine tag and purified by affinity chromatography. (B) Esterase activity of Ldh1p toward PNB. Km and Vmax values were calculated using Michaelis-Menten approximations. (C) TGL activity of Ldh1p toward DPG. (D) Purified Ldh1p and CRL were incubated on plates containing 2.5% trioleoylglycerol and 0.001% rhodamine B and, after 48 h, imaged at 350 nm. The numbers indicate the concentrations in mg/ml. In total, 200 l was loaded per agar slot. Hydrolysis of trioleoylglycerol was identified by fluorescent halos. RESULTS Enzymatic activity of Ldh1p. Characteristic GXSXG motifs and similarities to ␣/-hydrolases in the predicted protein sequences of Ldh1p suggest that the protein is an esterase or lipase (5, 28, 34). Indeed, Ldh1p was identified as a serine hydrolase by computational and chemical proteomics methods (2). We expressed Ldh1p as hexahistidine-tagged fusions in E. coli (Fig. 1A) and tested the isolated protein for esterase activity using PNB as a substrate. We found Ldh1p to be an active esterase hydrolyzing the model substrate PNB with a Km of 0.77 mM and a Vmax of 0.041 mol/min/mg (Fig. 1B). Phospholipase A, C, and D activities were not detected (data not shown). For the analysis of phospholipase A activity, we used the fluorogenic phospholipase A substrate bis-BODIPY FL C11-PC (B7701; Invitrogen) [1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-undecanoyl)-snglycero-3-phosphocholine]. For analysis of phospholipase C 778 DEBELYY ET AL. EUKARYOT. CELL FIG. 2. Hydrolase activity is not required for Ldh1p targeting to LDs. (A) Ldh1p is a hydrolytically active serine hydrolase with a classical catalytic triad containing a conserved serine (GXSXG motif), histidine, and aspartate (grey shading). (B) GFP-Ldh1m1p and Ldh1m1p-GFP were coexpressed in a yeast strain with genomically tagged Erg6p-RFP. Ldh1m1p colocalizes with the LD marker protein Erg6p [␦(24)-sterol methyl transferase]. Ldh1p containing a mutation of the active site (S177A) still localizes to LDs, indicating that the lipid targeting is independent of its catalytic activity. Bar, 1 m. BF, bright field. activity, we used the Amplex Red Phosphatidylcholine-Specific Phospholipase C Assay Kit (A12218; Invitrogen). Phospholipase D activity was assayed with the Amplex Red Phospholipase D Assay Kit (A12219; Invitrogen). Next, we assayed TGL activity using DPG as a substrate (Fig. 1C). One of the acyl residues of DPG contains the eximer-forming pyrene decanoic acid. Upon hydrolytic cleavage, the released pyrene decanoic acid leads to a decrease in eximer fluorescence. We found Ldh1p to be an active triacylglycerol lipase hydrolyzing the model substrate DPG with a Km of 3.3 mM and a Vmax of 1 mol/min/mg (Fig. 1C). TGL activity was also confirmed by an assay with fluorescein dilaurate as the substrate (not shown). We also adapted a specific and sensitive TGL assay originally developed for the measurement of bacterial TGLs (22). This assay is based on the hydrolysis of trioleoylglycerol and the formation of orange fluorescent rhodamine B halos. The results shown in Fig. 1D revealed that Ldh1p exerts a weak TGL activity. In summary, the purified Ldh1p exerts esterase and TG lipase activities. Mutational analysis of the GXSXG-type lipase motif. The characteristic GXSXG motif of ␣/-hydrolases is present in Ldh1p and is thought to contribute to the active site of the enzyme (Fig. 2A). To test this experimentally, we introduced a point mutation into the putative active site of Ldh1p (S177A) and analyzed the mutated protein for esterase activity. Replacement of serine with alanine in the hydrolase/lipase motif of Ldh1p completely abolished hydrolase activity. The mutated protein (Ldh1m1p) still localized to LDs, suggesting that the catalytic activity is not required for its topogenesis (Fig. 2B). The ⌬ldh1 mutant is characterized by the accumulation of lipids. Ldh1p has been shown to be predominantly localized to LDs (34a). To characterize the function of Ldh1p in more detail, we investigated whether the enzyme is involved in the biogenesis of LDs. To this end, LDs were isolated from oleic acid-induced wild-type and ⌬ldh1 mutant cells and appeared as a thick layer on top of a gradient of the mutant (Fig. 3A). The total weight of LDs was drastically increased in the ⌬ldh1 yeast strain in comparison to the wild type (Fig. 3B). These data were corroborated by TLC separation of extracted lipids from FIG. 3. The ⌬ldh1 yeast strain exhibits excessive accumulation of nonpolar and polar lipids in LDs during growth on medium containing oleic acid as a sole carbon source. (A) LDs were isolated from wildtype and ⌬ldh1 mutant cells and appeared as a thick layer on top of a gradient of the mutant. (B) The total weight of LDs was strongly increased in the ⌬ldh1 yeast strain in comparison to the wild type. The error bars indicate standard deviations. (C) TLC separation of extracted nonpolar and polar lipids from purified LDs, which showed the increase in nonpolar lipids and phospholipids in the ⌬ldh1 yeast strain. PC, phosphatidylcholine; PE, phosphatidylethanolamine; NPL, nonpolar lipids. VOL. 10, 2011 INVOLVEMENT OF YEAST HYDROLASE Ldh1p IN LIPID HOMEOSTASIS 779 wild-type cells, ⌬ldh1 mutant cells, and mutant cells expressing plasmids encoding either wild-type Ldh1p or the mutant Ldh1m1p. LDs appeared as a thick layer on top of the gradient, and comparison of the gradients revealed a thin lipid layer on top of the gradient for the wild type and the ⌬ldh1 mutant complemented with wild-type Ldh1p. A thicker layer, which is typical of the ⌬ldh1 mutant, was monitored for mutant cells that contained the catalytic dead Ldh1p (Fig. 5A). These data were corroborated by determination of the total weight of LDs, which was increased in the ⌬ldh1 strain and remained increased upon expression of the mutant protein (not shown). Accordingly, staining with Oil Red O and inspection of the cells by fluorescence microscopy (Fig. 5B), as well as by electron microscopy (Fig. 5C), revealed that the giant-LD phenotype of the ⌬ldh1 strain could be complemented with wild-type Ldh1p, but not with the catalytic dead mutant Ldh1p. These data demonstrate that functional complementation of the ⌬ldh1 mutant phenotype requires expression of enzymatically active Ldh1p, indicating that the hydrolase activity of the enzyme is required for its function in lipid homeostasis. DISCUSSION FIG. 4. Giant LDs in the ⌬ldh1 mutant. (A) Comparison of LD morphologies of the wild type (BY4742 Erg6p-RFP) and a deletion strain (BY4742 ⌬ldh1 Erg6p-RFP) by fluorescence microscopy. Bar, 1 m. (B) Localizations and morphologies of Oil Red O-stained wildtype (BY4742) and deletion strain (BY4742 ⌬ldh1) LDs. Bar, 1 m. (C) Absence of LDH1 leads to the formation of giant LDs, as well to the reduction of the total LD number in a cell. Shown are electron microscopic images of cells: the wild type (BY4742) and a deletion strain (BY4742 ⌬ldh1). Bars, 1 m. purified LDs, which showed the increase in nonpolar lipids and phospholipids in the ⌬ldh1 yeast strain (Fig. 3C). Giant lipid droplets in ⌬ldh1 mutant cells. To analyze whether the accumulation of lipids in mutant cells lacking the LD protein Ldh1p is accompanied by changes in LD morphology, the LDs of oleic acid-induced wild-type and ⌬ldh1 knockout cells expressing genomically encoded Erg6p-red fluorescent protein (RFP) were visualized by fluorescence microscopy (Fig. 4A), and the LDs of oleic acid-induced wild-type and ⌬ldh1 knockout cells were stained with Oil Red O and inspected by fluorescence microscopy (Fig. 4B). The data demonstrate that LDs can still be formed in the absence of Ldh1p, indicating that Ldh1p per se is not required for the formation of LDs. However, the morphological appearance of LDs in ⌬ldh1 mutant cells differed significantly from that in wild-type cells. The LDs of the mutant exhibited brighter fluorescence, indicating the existence of bigger LDs. These data were corroborated by electron microscopic inspection of wild-type and mutant cells, which revealed the presence of giant LDs in the ⌬ldh1 mutant (Fig. 4C). Esterase activity of Ldh1p is required for lipid homeostasis. The ⌬ldh1 yeast strain exhibits an excessive accumulation of lipids in LDs during growth on medium containing oleic acid as a sole carbon source. To test whether the loss of hydrolase activity of Ldh1p is responsible for the observed phenotype, we tested complementation of the mutant with functional and catalytic dead Ldh1p harboring a substitution of the active-site serine (Ldh1m1p). LDs were isolated from oleic acid-induced Ldh1p is a hydrolytically active serine hydrolase with a classical catalytic triad containing a serine (GXSXG motif). A conserved histidine was revealed by profile hidden Markov models (9a), and the aspartate of the probable triad was derived from an alignment with canine gastric triacylglycerol lipase (Fig. 2A). The putative active-site serine of Ldh1p is located next to the regions of highest hydrophobicity, suggesting that Ldh1p is a membrane-active hydrolase. We demonstrated that the hydrolase activity of Ldh1p could be completely abolished by the replacement of the active-site serine by alanine. Fluorescence microscopy analysis indicated that Ldh1p targets to the boundary of the LD monolayer membrane, supporting the idea that Ldh1p is involved in metabolic processes. Taken together, these features characterize Ldh1p as an active LD hydrolase. Mutation of the active site of Ldh1p does not lead to protein mislocalization, indicating that the lipase active site of Ldh1p is not involved in LD targeting. Cells deficient in Ldh1p are characterized by giant LDs accompanied by the accumulation of nonpolar lipids and phospholipids. Thus, Ldh1p seems to be required for the mobilization of LD-stored lipids, which would also explain the dependency of the Ldh1p function on its hydrolase activity. We speculate that Ldh1p plays a role in maintaining lipid homeostasis by regulating both phospholipid and nonpolar lipid levels. Interestingly, the ⌬ldh1 (⌬ybr204c) strain has been reported to exhibit resistance to the lipophilic drug camptothecin (16, 19, 20). Camptothecin is a cytotoxic quinoline alkaloid that inhibits the DNA enzyme topoisomerase I. The resistance to camptothecin might be explained by increased detoxification properties of LDs with an excessive amount of nonpolar lipids, which may serve as a reservoir for hydrophobic toxic molecules (3, 7, 21, 35). Global genomic screening research recently disclosed the transient induction of LDH1 by growth on oleate medium (30). It was shown that the level of Ldh1p increased within the first 3 h of induction, followed by a decrease within the subsequent 6 h and complete reduction to basal levels within the next 17 h. Such an expression profile might hint at a 780 EUKARYOT. CELL DEBELYY ET AL. FIG. 5. The esterase activity of Ldh1p is required for lipid homeostasis. The ⌬ldh1 yeast strain exhibits excessive accumulation of lipids in LDs during growth on medium containing oleic acid as a sole carbon source. LDs were isolated from oleic acid-induced wild-type cells and ⌬ldh1 mutant cells expressing plasmids encoding either wild-type Ldh1p or the mutant Ldh1m1p. (A) LDs appeared as a thick layer on top of the gradient, and comparison of the gradients revealed a thin lipid layer on top of the gradient for the wild type (WT) and the ⌬ldh1 mutant complemented with wild-type Ldh1p. A thicker layer, typical of the ⌬ldh1 mutant, was monitored for mutant cells that contained the catalytic dead Ldh1p. (B) Staining with Oil Red O and inspection by fluorescence microscopy revealed that the giant-LD phenotype of the ⌬ldh1 strain could be complemented with wild-type Ldh1p, but not with the catalytic dead mutant Ldh1p. Bar, 1 m. (C) Electron microscopy revealed that the giant-LD phenotype of the ⌬ldh1 strain could be complemented with wild-type Ldh1p, but not with the catalytic dead mutant Ldh1p. Bar, 1 m. regulatory or signaling function instead of direct involvement of the enzyme in lipid metabolism. Interestingly, LDH1 expression is also induced upon sporulation (6), which is mildly affected in cells deficient in Ldh1p (25). Our data clearly show that Ldh1p per se is not required for the biogenesis of LDs, but the severe accumulation of lipids and the corresponding appearance of the giant LDs in ⌬ldh1 mutant cells strongly suggest a role for the enzyme in LD lipid homeostasis. ACKNOWLEDGMENTS We thank Elisabeth Becker, Monika Bürger, and Uta Ricken for technical assistance; Robert Rucktäschel for scientific input; and Wolfgang Girzalsky for reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB642 and ER178/4-1). REFERENCES 1. Altmann, R. 1890. Die Elementarorganisem und ihre Beziehungen zu den Zellen. Veit, Leipzig, Germany. 2. Baxter, S. M., et al. 2004. Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Mol. Cell Proteomics 3:209–225. 3. Beller, M., K. Thiel, P. J. Thul, and H. Jackle. 2010. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 584:2176–2182. 4. Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 5. Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528–530. 6. Chu, S., et al. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705. VOL. 10, 2011 INVOLVEMENT OF YEAST HYDROLASE Ldh1p IN LIPID HOMEOSTASIS 7. Czabany, T., K. Athenstaedt, and G. Daum. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771:299–309. 8. Daum, G., P. C. Bohni, and G. Schatz. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028– 13033. 9. Digel, M., R. Ehehalt, and J. Fullekrug. 2010. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 584:2168–2175. 9a. Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. 10. Erdmann, R., M. Veenhuis, D. Mertens, and W. H. Kunau. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86:5419–5423. 11. Erdmann, R., et al. 1991. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64:499–510. 12. Farese, R. V., Jr., and T. C. Walther. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139:855–860. 13. Fujimoto, T., Y. Ohsaki, J. Cheng, M. Suzuki, and Y. Shinohara. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130:263– 279. 14. Goodman, J. M. 2008. The gregarious lipid droplet. J. Biol. Chem. 283: 28005–28009. 15. Guo, Y., K. R. Cordes, R. V. Farese, Jr., and T. C. Walther. 2009. Lipid droplets at a glance. J. Cell Sci. 122:749–752. 16. Hsiang, Y. H., R. Hertzberg, S. Hecht, and L. F. Liu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260:14873–14878. 17. Imanishi, Y., V. Gerke, and K. Palczewski. 2004. Retinosomes: new insights into intracellular managing of hydrophobic substances in lipid bodies. J. Cell Biol. 166:447–453. 18. Ingelmo-Torres, M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic 10:1785–1801. 19. Knab, A. M., J. Fertala, and M. A. Bjornsti. 1993. Mechanisms of camptothecin resistance in yeast DNA topoisomerase I mutants. J. Biol. Chem. 268:22322–22330. 20. Knab, A. M., J. Fertala, and M. A. Bjornsti. 1995. A camptothecin-resistant DNA topoisomerase I mutant exhibits altered sensitivities to other DNA topoisomerase poisons. J. Biol. Chem. 270:6141–6148. 21. Kohlwein, S. D. 2010. Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285:15663–15667. 22. Kouker, G., and K. E. Jaeger. 1987. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 53:211–213. 781 23. Krahmer, N., Y. Guo, R. V. Farese, Jr., and T. C. Walther. 2009. SnapShot: lipid droplets. Cell 139:1024–1024.e1021. 24. Listenberger, L. L., and D. A. Brown. 2008. Lipid droplets. Curr. Biol. 18:R237–R238. 25. Marston, A. L., W. H. Tham, H. Shah, and A. Amon. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367– 1370. 26. Martin, S., and R. G. Parton. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378. 27. Mlícková, K., et al. 2004. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 70:3918–3924. 28. Schrag, J. D., and M. Cygler. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 284:85–107. 29. Skinner, J. R., et al. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284:30941–30948. 30. Smith, J. J., et al. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259–271. 31. Stobart, A. K., S. Stymne, and S. Höglund. 1986. Safflower microsomes catalyse oil accumulation in vitro: a model system. Planta 169:33–37. 32. Tauchi-Sato, K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507–44512. 33. Thiele, C., and J. Spandl. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20:378–385. 34. Thoms, S., M. O. Debelyy, K. Nau, H. E. Meyer, and R. Erdmann. 2008. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 275:504–514. 34a.Thoms, S., M. O. Debelyy, M. Connerth, G. Daum, and R. Erdmann. 2011. The putative Saccharomyces cerevisiae hydrolase Ldh1p is localized to lipid droplets. Eukaryot. Cell 10:●. 35. Walther, T. C., and R. V. Farese, Jr. 2009. The life of lipid droplets. Biochim. Biophys. Acta 1791:459–466. 36. Welte, M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17:363–369. 37. Wilson, E. 1896. The cell in development and inheritance. Macmillan, New York, NY. 38. Zweytick, D., K. Athenstaedt, and G. Daum. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta 1469:101–120. CHAPTER 3. DISCUSSION_________________________________________________ 77 CHAPTER 3. DISCUSSION The novel hydrolases and peroxisome related ubiquitin-specific protease of yeast S. cerevisiae are characterized in this chapter. Lpx1p and Ldh1p are hydrolases of peroxisome and lipid droplets, respectively. Ubp15p is a peroxisome related deubiquitinating enzyme. Triacylglycerol lipase and hydrolase activities were shown for both recombinant proteins Lpx1p and Ldh1p as well as oligoubiqutin-hydrolase and Ub-Pex5p deubiquitinating activities were shown for recombinant Ubp15p in vitro. It is demonstrated that the Lpx1p protein is not required for wild-type-like steady-state function of peroxisomes and that Δlpx1 mutants have an aberrant morphology characterized by intraperoxisomal vesicles or invaginations. Morover, Ldh1p is not required for the function and biogenesis of peroxisomes, but is essential for the maintenance of a steady-state level of the nonpolar and polar lipids of lipid droplets. In line with this finding, the Δldh1 strain is characterized by appearance of giant lipid droplets and an excessive accumulation of nonpolar lipids and phospholipids upon growth on medium containing oleic acid as a sole carbon source. It is demonstrated that the peroxisomal AAA-complex contains Pex5p dislocase and Ub-Pex5p deubiquitinating activites and that Ubp15p is a novel constituent of this complex. Δubp15 mutant is characterized as a strain which has a stress related PTS1import defect. 3.1 Novel hydrolases of yeast S. cerevisiae Lpx1p as well as Ldh1p, a novel hydrolase of S. cerevisiae (35, 209), comprises the typical GXSXG-type lipase motif of members of the α/β-hydrolase family (189). LPX1 is one of the most strongly induced genes following a shift from glucose to oleate, as determined by serial analysis of gene expression (SAGE) experiments (103). The oleate-induced increase in mRNA abundance is abolished in the Δpip2 Δoaf1 double deletion strain, indicating that its induction is dependent on the transcription factor pair Pip2p and Oaf1p (103). It was shown by use of an antibody raised against Lpx1p that this protein itself is induced by oleic acid (210). Besides, it was determined, by using a Protein A tag, that Lpx1p protein is strongly induced by oleic acid (196). Moderate induction by oleic acid was also demonstrated for Ldh1p protein (196). CHAPTER 3. DISCUSSION_________________________________________________ 78 Both proteins Lpx1p as well as Ldh1p carry a putative peroxisomal targeting signal type-1 (PTS1) (126) and can be aligned with two regions of homology by WUBLAST-2 search (134) (Fig. 3.1.1). Fig. 3.1.1 Ldh1p and Lpx1p from S. cerevisiae are similar proteins with a α/βhydrolase/lipase motif. Alignment of the two regions of homology of Lpx1p and Ldh1p exhibiting 28% (region A) and 27% (region B) amino acid identities. The GXSXG hydrolase/lipase motif is underlined; similar amino acids are indicated by a plus symbol. Taken with modifications from (209). Lpx1p does not conform with its QKL motife to the general PTS1 consensus. Three other proteins with an QKL on their extreme C-terminus are known in S. cerevisiae, which are probably not peroxisomal: Efb1p (systematic name: Yal003wp) is the elongation factor EF-1b (82), Rpt4p (Yor259cp) is a mostly nuclear 19S proteasome cap AAA protein (149), and Tea1p (Yor337wp) is a nuclear Ty1 enhancer activator (70). However, QKL is sufficient to sponsor Pex5p binding (124). Why are these QKL proteins not imported into peroxisomes? This is probably due to the upstream sequences. Lpx1p has a lysine at position -1 (relative to the PTS1 tripeptide) and a hydrophobic amino acid at position -5. These features promote Pex5p binding and are not found in the other three QKL proteins (124). Ldh1p contains the consensus sequence for a classical peroxisomal targeting signal type-1 (PTS1), but the protein is primarily targeted to lipid droplets and not to peroxisomes. Peroxisomal exclusion of Ldh1p is likely due to the upstream sequences with charged amino acids in positions - 2 and - 5. These positions are adverse to Pex5p binding and peroxisomal localization, for which polar/hydrophilic or positively charged amino acids in position - 2 are preferred. The negatively charged amino acid is not even counteracted by neighbouring amino acids, giving the likely explanation for dominating peroxisomal exclusion. The classical PTS1, SKL, is not completely sufficient to target protein to peroxisomes if the upstream sequences are not supportive. It was shown that the majority of Ldh1p is a lipid droplets protein that is targeted independently of the PTS1-binding Pex5p. CHAPTER 3. DISCUSSION_________________________________________________ 79 Moreover, it was shown by applying a PTS1 prediction algorithm (http://mendel.imp.ac.at/pts1/) (156, 157), which predicted peroxisomal localization, that only Lpx1p but not Ldh1p as well as Efb1p, Rpt4p, and Tea1p, is localized in peroxisome. It was shown dimerization of Lpx1p in the context of piggyback protein import into peroxisomes (210). Self-interaction (dimerization) is frequently found in regulation of the enzymatic activity of other lipases such as Candida rugosa lipase or human lipoprotein lipase (63, 164). Lipid droplets localization signals are only poorly characterized. It has been suggested that lipid droplets localization signals are constituted of hydrophobic residues at the Cterminus of a protein (154, 237). A Kyte-Doolittle plot of Ldh1p indicated a region with particularly high hydrophobicity from amino acids 130 to 154. This stretch might be required to target and/or to attach Ldh1p to lipid droplets. Indeed, it was shown that lipid droplets targeting are not abrogated when GFP is added to the C-terminus or the N-terminus of Ldh1p. Thus, targeting information within central parts of Ldh1p, rather than at its termini, is sufficient for the lipid droplets localization. Interestingly, the Lpx1p stretch of high hydrophobicity is in a similar location in the primary sequence, namely, at amino acids 154 to 177 (210). The hydrophobic stretches in Ldh1p are likely not classical transmembrane domains, because lipid droplets are bound by a single monolayer membrane of phospholipids. Extended localization studies of Ldh1p-GFP showed that at least a portion of the polypeptide is targeted to peroxisomes and mitochondria. While this triple localization may reflect the true cellular scenario, it has to be taken into account that partial targeting of Ldh1p to peroxisomes and mitochondria may be due to the overexpression of Ldh1p-GFP. The putative active-site serine of Lpx1p is located next to the region of highest hydropathy, suggesting that Lpx1p is a membrane-active lipase that contributes to metabolism or the membrane shaping of peroxisomes. Peroxisomes are sites of lipid metabolism (223). It is thus not surprising to find a lipase associated with peroxisomes. It was demonstrated that Lpx1p has triacylglycerol lipase activity; however, activities towards the artificial test substrates DPG (1,2-dioleoyl-3-(pyren-1-yl) decanoyl-rac-glycerol) and DGR (1,2-O-dilaurylrac-glycero-3-glutaric acid (6-methyl resorufin) ester) were low (210). The evidence for phospholipase A activity of the enzyme (substrate: 1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora3a,4a-diaza-sindacene-3-undecanoyl)-sn-glycero-3-phosphocholine), together with the electron microscopy phenotype, suggest that Lpx1p has a more specialized role in modifying membrane phospholipids (210). A mammalian group VIB calcium-independent phospholipase A2 (iPLA2c) was identified that possesses a PTS1 SKL and a mitochondrial CHAPTER 3. DISCUSSION_________________________________________________ 80 targeting signal (140, 234). The enzyme is localized in peroxisomes and mitochondria, and is involved, among others, in arachidonic acid and cardiolipin metabolism (139, 155). Knockout mice of iPLA2c show mitochondrial ⁄ cardiac phenotypes (141). It will be exciting to determine whether human iPLA2c and yeast Lpx1p are functionally related. It was shown that peroxisomes are still functional in the absence of LPX1. This suggests a non-essential metabolic role for Lpx1p in peroxisome function (210). The morphological defect found in electron microscopic images of a deletion of Lpx1p (peroxisomes containing inclusions or invaginations) is symptomatic of a yeast peroxisomal mutant, and is reminiscent of the phenotypes found in human peroxisomal disorders (56, 151). All these data suggest that Lpx1p is required to determine the shape of peroxisomes (210). Lipase activity and cellular function of Lpx1p could be involved in various processes: (a) detoxification and stress response, (b) lipid mobilization, or (c) peroxisome biogenesis. As Lpx1p expression may be regulated by Yrm1p and Yrr1p (135), a transcription factor pair that mediates pleiotropic drug resistance effects, it was speculated that Lpx1p is required for a multidrug resistance response (210). The epoxide hydrolase activity for Lpx1p was, however, excluded because hydrolysis of the epoxide hydrolase test substrate was not affected by a specific epoxide hydrolase inhibitor (210). It was demonstrated that recombinant Ldh1p exerts an esterase and triacylglycerol lipase activities. The enzyme activity was abolished upon mutation of the conserved GXSXGtype lipase motif of the protein. The S. cerevisiae Δldh1 strain is characterized by the appearance of giant lipid droplets and the accumulation of nonpolar lipids and phospholipids in lipid droplets, indicative of a role of Ldh1p in maintaining lipid homeostasis (35). Ldh1p is a hydrolytically active serine hydrolase with a classical catalytic triad containing a serine (GXSXG motif). A conserved histidine was revealed by profile hidden Markov models (40), and the aspartate of the probable triad was derived from an alignment with canine gastric triacylglycerol lipase (209). The putative active-site serine of Ldh1p is located next to the regions of highest hydrophobicity, suggesting that Ldh1p is a membrane-active hydrolase. It was demonstrated that the hydrolase activity of Ldh1p could be completely abolished by the replacement of the active-site serine by alanine. Fluorescence microscopy analysis indicated that Ldh1p targets to the boundary of the lipid droplets monolayer membrane, supporting the idea that Ldh1p is involved in metabolic processes. Taken together, these features characterize Ldh1p as an active lipid droplets hydrolase. Mutation of the active site of Ldh1p does not lead to protein mislocalization, indicating that the lipase active site of Ldh1p is not involved in lipid droplets targeting. CHAPTER 3. DISCUSSION_________________________________________________ 81 Cells deficient in Ldh1p are characterized by giant lipid droplets accompanied by the accumulation of nonpolar lipids and phospholipids. Thus, Ldh1p seems to be required for the mobilization of lipid droplets-stored lipids, which would also explain the dependency of the Ldh1p function on its hydrolase activity. It was speculated that Ldh1p plays a role in maintaining lipid homeostasis by regulating both phospholipid and nonpolar lipid levels (35). Interestingly, the Δldh1 (Δybr204c) strain has been reported to exhibit resistance to the lipophilic drug camptothecin (89, 110, 111). Camptothecin is a cytotoxic quinoline alkaloid that inhibits the DNA enzyme topoisomerase I. The resistance to camptothecin might be explained by increased detoxification properties of lipid droplets with an excessive amount of nonpolar lipids, which may serve as a reservoir for hydrophobic toxic molecules (15, 32, 112, 221). Global genomic screening research recently disclosed the transient induction of LDH1 by growth on oleate medium (196). It was shown that the level of Ldh1p increased within the first 3 h of induction, followed by a decrease within the subsequent 6 h and complete reduction to basal levels within the next 17 h. Such an expression profile might hint at a regulatory or signalling function instead of direct involvement of the enzyme in lipid metabolism. Interestingly, LDH1 expression is also induced upon sporulation (29), which is mildly affected in cells deficient in Ldh1p (142). The data clearly show that Ldh1p per se is not required for the biogenesis of lipid droplets, but the severe accumulation of lipids and the corresponding appearance of the giant LDs in Δldh1 mutant cells strongly suggest a role for the enzyme in lipid droplets lipid homeostasis (35). While Lpx1p was shown to be a peroxisomal enzyme, subcellular localization studies revealed that Ldh1p is predominantly localized to lipid droplets. It was shown that Lpx1p import is dependent on the PTS1 receptor Pex5p. Moreover, it was shown that Lpx1p is piggyback-transported into peroxisomes. But it was demonstrated that targeting of Ldh1p to lipid droplets occurs independently of the PTS1 receptor Pex5p. Triacylglycerol lipase as well as hydrolase activities were shown for both recombinant proteins Lpx1p and Ldh1p in vitro. It was shown that the Lpx1p protein is not required for wild-type-like steadystate function of peroxisomes, which might be indicative of a metabolic rather than a biogenetic role. It was clearly shown that peroxisomes in Δlpx1 mutants have an aberrant morphology characterized by intraperoxisomal vesicles or invaginations. It was demonstrated that Ldh1p is not required for the function and biogenesis of peroxisomes. Ldh1p is required for the maintenance of a steady-state level of the nonpolar CHAPTER 3. DISCUSSION_________________________________________________ 82 and polar lipids of lipid droplets. A characteristic feature of the Δldh1 strain is the appearance of giant lipid droplets and an excessive accumulation of nonpolar lipids and phospholipids upon growth on medium containing oleic acid as a sole carbon source. Ldh1p is thought to play a role in maintaining the lipid homeostasis in yeast by regulating both phospholipid and nonpolar lipid levels. Ldh1p hydrolase and the Lpx1p lipase are not redundant proteins; other enzymes, probably with somewhat lower homology, cannot compensate for a defect in the two enzymes. Both peroxisomes and lipid droplets function in concert in lipid metabolism. Lipid droplets require the action of triacylglycerol lipases to metabolize nonpolar lipids, while peroxisomes represent the sole cellular site for fatty acid oxidation. It is thus possible that the peroxisomal Lpx1p and the lipid droplets Ldh1p play a physiological role in lipid metabolism by mobilizing fatty acids and channeling them to their site of degradation. Lipid droplets, as fatty acid depot organelles, can be the storage sites for nonpolar lipids that are further metabolized in peroxisomes. For this reason, and not surprisingly, LDs have been found in proximity to peroxisomes in different organisms (14, 78, 187). It was also shown that S. cerevisiae peroxisomes attach to lipid droplets or even project into lipid droplets, which was interpreted as an intimate interaction between the two compartments (18). Ldh1p and Lpx1p shows that beyond a metabolic collaboration, peroxisomes and lipid droplets may be equipped with similar hydrolases (209). 3.2 Ubp15p, a novel compound of AAA-complex It was proposed, at least for Pex1p, that it can fulfil its function by unfoldase activity, using its N-terminal putative adaptor-binding domain (193). So far, it is possible only speculate, that both AAA peroxins, Pex1p and Pex6p are highly substrate specific unfoldases/foldase (chaperons), enzymes that unfold/fold protein substrate in ATP-hydrolysis dependent manner. In this case, ubiquitinated Pex5p could be a substrate for such activity. On the first step of the Ub-Pex5p release from the peroxisome it could be unfolded by one of AAA peroxin, probably by Pex6p. On the next step, Pex5p could be folded by second AAA peroxin Pex1p, with the following release of Pex5p to the cytosol. Such hypothesis could explain a requirement of two AAA ATPases, instead of just one. It was recently shown that the AAA-complex is responsible for receptor deubiquitination, which is supposed to be an important step in receptor recycling (34). CHAPTER 3. DISCUSSION_________________________________________________ 83 It was shown that yeast S. cerevisiae has 19 deubiquitinating enzymes (DUB) (Table 3.2.1). All of them have catalytic regions with three evolutionary conserved amino acids: cysteine, hystedine and tryptophan (Fig 3.2.1). Fig. 3.2.1 Highest homology region of S. cerevisiae deubiquitinating enzymes. Moderately conserved residues are shaded in grey whereas the conserved histidine (H) and tryptophan (D), two aminoacids of the catalytic triad CHD, as well as highly conserved tyrosine (Y) are highlighted in black. ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) algorithm was used for yeast S. cerevisiae DUBs alignment. The corresponding deubiquitinating enzymes (DUB) Ubp15p was identified as a novel factor that accompanies the AAA-complex in peroxisomal protein import (34). It was demonstrated that Ubp15p share common features conserved among other UBP members (90) (Fig 3.2.2). All members of the ubiquitin-specific processing protease family (UBP) of deubiquitinating enzymes (DUB) share strong homology in the Cys and His Boxes. The Cys Box contains the catalytic cysteine residue, which is thought to undergo deprotonation and to unleash a nucleophilic attack on the carbonyl carbon atom of the ubiquitin Gly76 at the scissile peptide bond. In analogy with other cysteine proteases, the deprotonation of this cysteine residue most likely is assisted by an adjacent His residue, which, in turn, is stabilized by a nearby side chain from an Asn or Asp residue. Together, these three residues constitute the so-called catalytic triad (90). Previous mutagenesis studies on several UBPs have provided evidence that these residues have critical roles in catalysis (9, 61, 90, 91). PolyUb-Pex5p (106, 171) as well as monoUb-Pex5p (71, 116) are solely found at the peroxisomal membrane fraction in wild-type yeast and rat liver cells, indicating that Pex5p ubiquitination exclusively takes place at the peroxisomal membrane. Interestingly, exported Pex5p appears to be unmodified, indicating that the Ub-moiety is removed during or directly after receptor export (27, 116, 169). However, published data on the deubiquitination of Pex5p so far have focused on in vitro assays with mammalian Pex5p. CHAPTER 3. DISCUSSION_________________________________________________ 84 Table 3.2.1 Deubiquitinating enzymes of yeast Saccharomyces cerevisiae DUB 1 Ubp1p 2 Ubp2p 3 Ubp3p 4 Doa4p 5 Ubp5p 6 Ubp6p 7 Ubp7p 8 Ubp8p 9 Ubp9p 10 Ubp10p 11 Ubp11p 12 Ubp12p 13 Ubp13p 14 Ubp14p 15 Ubp15p 16 Ubp16p 17 Yuh1p 18 Otu1p 19 Rpn11p Description Cellular component Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins; cleaves at the C terminus of ubiquitin fusions irrespective of their size; capable of cleaving polyubiquitin chains. Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins; interacts with Rsp5p and is required for MVB sorting of membrane proteins; can cleave polyubiquitin and has isopeptidase activity. Ubiquitin-specific protease that interacts with Bre5p to coregulate anterograde and retrograde transport between the ER and Golgi; inhibitor of gene silencing; cleaves ubiquitin fusions but not polyubiquitin; also has mRNA binding activity. Ubiquitin isopeptidase, required for recycling ubiquitin from proteasome-bound ubiquitinated intermediates, acts at the late endosome/prevacuolar compartment to recover ubiquitin from ubiquitinated membrane proteins en route to the vacuole. cytoplasm, endoplasmic reticulum (1, 4, 12, 23, 34, 77, 109, 115, 125, 130, 133, 214) cytoplasm (12, 23, 122, 125) cytoplasm (12, 30, 31, 148) endosome, membrane fraction, proteasome complex, mitochondrion (1, 3, 5, 6, 109, 161, 162, 174, 202) Putative ubiquitin-specific protease, closest paralog of Doa4p but has no functional overlap; concentrates at the bud neck. cellular bud neck, incipient cellular bud site proteasome complex, proteasome regulatory particle (3, 5, 115, 161) cytoplasm (5, 83) Ubiquitin-specific protease that is a component of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) acetylation complex; required for SAGA-mediated deubiquitination of histone H2B. Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions. DUBm complex, SAGA complex, SLIK (SAGA-like) complex cytoplasm (5, 79, 83) Ubiquitin-specific protease that deubiquitinates ubiquitin-protein moieties; may regulate silencing by acting on Sir4p; involved in posttranscriptionally regulating Gap1p and possibly other transporters; primarily located in the nucleus. Ubiquitin-specific protease that cleaves ubiquitin from ubiquitinated proteins. nucleus (5, 101, 102, 194) UNKNOWN (5, 125) Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease present in the nucleus and cytoplasm that cleaves ubiquitin from ubiquitinated proteins. Putative ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions. cytoplasm, nucleus (5, 23, 92) UNKNOWN (5, 77, 83) Ubiquitin-specific protease that specifically disassembles unanchored ubiquitin chains; involved in fructose-1,6bisphosphatase (Fbp1p) degradation; similar to human isopeptidase T. Ubiquitin-specific protease that may play a role in ubiquitin precursor processing. cytoplasm (1, 4, 130) cytoplasm, peroxisome (1, 23, 34, 115, 133) Deubiquitinating enzyme anchored to the outer mitochondrial membrane, probably not important for general mitochondrial functioning, but may perform a more specialized function at mitochondria. Ubiquitin C-terminal hydrolase that cleaves ubiquitin-protein fusions to generate monomeric ubiquitin; hydrolyzes the peptide bond at the C-terminus of ubiquitin; also the major processing enzyme for the ubiquitin-like protein Rub1p. Deubiquitylation enzyme that binds to the chaperone-ATPase Cdc48p; may contribute to regulation of protein degradation by deubiquitylating substrates that have been ubiquitylated by Ufd2p; member of the Ovarian Tumor (OTU) family. Metalloprotease subunit of the 19S regulatory particle of the 26S proteasome lid; couples the deubiquitination and degradation of proteasome substrates; involved, independent of catalytic activity, in fission of mitochondria and peroxisomes. cytoplasm, mitochondrial outer membrane (109, 133) cytoplasm (12, 23, 125, 131, 182, 214) cytoplasm, nucleus (22, 59, 98, 137, 181) cytosol, mitochondrion, nucleus, proteasome regulatory particle, lid subcomplex, proteasome storage granule (16, 74, 85, 220, 235) Ubiquitin-specific protease situated in the base subcomplex of the 26S proteasome, releases free ubiquitin from branched polyubiquitin chains; works in opposition to Hul5p polyubiquitin elongation activity; mutant has aneuploidy tolerance. Ubiquitin-specific protease that cleaves ubiquitin-protein fusions. References (1, 23, 74) (77, 115) CHAPTER 3. DISCUSSION_________________________________________________ 85 Fig 3.2.2 Sequence alignment of Ubp15p with six representative UBP family proteins Conserved residues are shaded in yellow whereas the catalytic triad is highlighted in red. Residues that are involved in direct inter-molecular hydrogen bond interactions using their side chains and main chains are marked with purple and green arrows, respectively. Residues that are involved in van der Waals contact with ubiquitin aldehyde (Ubal) are labelled with blue squares. Residues that coordinate the oxyanion through hydrogen bonds are identified with blue triangles above the alignment. The secondary structural elements above the sequences are indicated for the free HAUSP (lower) and the ubiquitin-bound HAUSP (upper), respectively. Taken with modifications from (90). CHAPTER 3. DISCUSSION_________________________________________________ 86 Soluble monoUb-Pex5p is formed when the in vitro export reaction is performed in presence of DUB inhibitors (27, 71). Accordingly, it was concluded that deubiquitination of Pex5p occurs predominantly in the cytosol after release from the membrane. It also was suggested that a small fraction of the dislocated Ub-Pex5p in vitro can already be deubiquitinated by reducing reagents like glutathione, while most of the Ub-Pex5p is deubiquitinated via an enzymatic pathway (71). Cleavage of the Ub-moiety from mammalian Pex5p was originally thought to be catalyzed by an unspecific reaction that could be carried out by any DUB in the cytosol or may even function via a non-enzymatic reaction (71). Later it was shown that deubiquitination of yeast Pex5p represents a specific and important event for the optimal functionality of the export machinery (34). Ubp15p has been identified as deubiquitinating enzyme that is dedicated for this deubiquitination event in baker’s yeast. The deubiquitinating activity found to be associated with the endogenous AAA-complex was the first indication for the presence of such an enzyme. Mass spectrometry analysis of the AAA-complex derived from endogenous proteins as well as overexpressed Pex6p revealed a stable association of Ubp15p. The interaction with Pex6p was confirmed by yeast two-hybrid analysis and the interaction site could be mapped to the D1 domain of Pex6p. While the evolutionarily related AAA-protein Cdc48p (p97/VCP) utilizes several co-factors (98), Ubp15p is only the second known co-factor that accompanies the function of Pex6p, with its membrane-anchor Pex15p (Pex26p in mammals) being the first one (20). Pex6p acts in concert with Pex1p as dislocase complex for the ubiquitinated Pex5p in order to facilitate the export of the PTS1-receptor back to the cytosol (150, 172). This leads to the intriguing question, how the activity of the deubiquitinating enzyme Ubp15p is coordinated with the Ub-dependent dislocation of Pex5p from the membrane and release into the cytosol. The finding that the deletion of UBP15 does not result in a complete peroxisomal biogenesis defect, can either be explained by the model that deubiquitination has only modulating activity or it may indicate the existence of additional factors which may accompany the AAA-complex in its function. This situation could well be explained by redundant DUBs acting on Ub-Pex5p. Possible candidates are the known Ubp15p-binding partners Ubp14p and Doa4p (Ubp4p) (4, 120). However, the characterization of the single deletion strains suggested that these two DUBs do not have a peroxisome-specific function similar to Ubp15p. The single deletion strain of Ubp14p had no significant effect on peroxisome morphology or cargo import, both under oleate as well as under H2O2 stress conditions. Previous studies have CHAPTER 3. DISCUSSION_________________________________________________ 87 suggested a role for Ubp14p in the disassembly of unanchored polyubiquitin chains (4). The deletion of Doa4p had an effect on the efficiency of peroxisomal cargo import. However, it has to be taken into account that the deletion of Doa4p is known to result in pleiotropic effects on many Ub-dependent processes in the cell, as Doa4p influences the homeostasis of free ubiquitin (202). Possibly related to this function, DOA4 is a stress regulated gene, giving an alternative explanation for the oleate induction reported by (197). Thus, although it is not possible to fully exclude that Doa4p exhibits a peroxisome-related overlapping function with Ubp15p, the partial import defect observed for the Δdoa4 strain might well be explained by the pleiotropic phenotype of this mutant. The observation that Δubp15 cells contain more clustered peroxisomes than wild-type cells is puzzling. Earlier work correlated a reduced level of imported matrix proteins such as catalase and the occurrence of clustered peroxisomes (238). Slowing of Pex5p cycling is most likely associated with reduced import rates. Interestingly, induction of oxidative stress by treating cells with hydrogen peroxide causes Pex5p to amass on the organelle membrane and significantly reduces PTS1 protein import (127, 165, 206). As data are clear in that Ubp15p can deubiquitinate Pex5p and as the ubiquitination status of the PTS1-receptor directly influences its cycling (27, 169), it is conceivable that the deletion of Ubp15p influences the import process of PTS1-proteins like catalase and thus possibly also morphology and clustering of peroxisomes. Although Ubp15p is not essential for peroxisomal biogenesis under normal conditions, its regulative function gains significantly more weight when the cells are stressed with H2O2 and require an efficient import of matrix proteins into peroxisomes. Thus, the findings that 1) Ubp15p is stably associated with the export machinery by interacting with Pex6p, 2) the fact that a small portion of the protein is associated with peroxisomes, and 3) the partial protein import defect for PTS1 proteins observed in Δubp15 cells upon oxidative stress suggest that the deubiquitination, at least in baker’s yeast, is not an unspecific event that takes place at any location in the cytosol, as suggested by the mammalian study (71), but supports the notion that the detachment of the Ub-moiety is a regulated event. Ubiquitination of the receptor is a precondition for its export (27, 169). In this respect, it is likely that the Pex1p/Pex6p-complex recognizes the Ub-moiety. This, however, still needs to be shown. The in vitro data demonstrate that the exported import receptor is deubiquitinated. This reflects the in vivo situation which is clear in that the cytosolic receptor is not ubiquitinated. Thus, the accumulating evidence indicates that the ubiquitin moiety is cleaved off from the import receptor during or shortly after export. CHAPTER 3. DISCUSSION_________________________________________________ 88 There are several possible advantages to favour a peroxisome-associated deubiquitination of Ub-Pex5p. This could protect Pex5p from unspecific ubiquitination by detaching Ub-moieties from lysine residues or preventing the formation of a poly-ubiquitin chain at the crucial cysteine residue dedicated to mono-ubiquitination (Fig 3.2.3). This function would ensure an optimal protection and presentation of monoUb-Pex5p to the export machinery Fig 3.2.3 The PTS1-receptor cycle. Hypothetical model describes a possible function of Ubp15p in Pex5p cycle. Ubp15p can protect Pex5p from unspecific polyubiquitination by ubiquitin-conjugating enzyme (E2) Ubc4p. Such activity of Ubp15p can prevent Pex5p proteasomal degradation and save it for the next round of recetor cycle. Red colored arrows show direction of the Pex5p cycle with participation of Ubp15p; blue colored arrow show direction of the Pex5p cycle without participation of Ubp15p. DUB, unknown deubiquitinating enzyme; P, proteasome; Ub, ubiquitin. Another possible explanation might be that the deubiquitination step may trigger the efficient release of Pex5p from the export-machinery by cleavage of the complex-bound Ubmoiety. Furthermore, this mechanism could prevent the monoUb-Pex5p to be recognized by the proteasome system to ensure an efficient recycling of the receptor for new matrix protein import cycles. The finding that both ubiquitinating and deubiquitinating activities are required for the transport of proteins from a membrane to the cytosol finds an examples in the ERAD CHAPTER 3. DISCUSSION_________________________________________________ 89 pathway. The AAA-type ATPase p97 (Cdc48/VCP) is evolutionary related to the peroxisomal AAA-proteins Pex1p and Pex6p (58). Most interestingly, among the growing number of known co-factors and adaptor proteins that p97 utilizes to carry out its different functions are also several deubiquitinating enzymes (98). The mammalian deubiquitinating enzymes YOD1 and Ataxin-3 are p97-associated proteins and function in the ERAD pathway (39, 45, 224). Most of the published literature defines both DUBs as a positive regulator of the p97driven dislocation of the ERAD-substrates, most likely by editing the poly-Ub chains on the substrates themselves in order to ensure the best fit to downstream Ub-receptor proteins. Ubp15p acts in concert with the AAA-peroxins in the matrix protein import cycle of the PTS1-receptor. Pex5p deubiquitination occurs as a highly specific event in yeast and removal of ubiquitin of the PTS1-receptor Pex5p turns out to be a vital step in the receptor cycle in its own right. Thus, removal of the ubiquitin seems to complete the receptor cycle of Pex5p in order to make the receptor available for another round of matrix protein import. CHAPTER 4. REFERENCES________________________________________________ 90 CHAPTER 4. REFERENCES 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. Aguilar-Henonin, L., J. Bravo, and P. Guzman. 2006. Genetic interactions of a putative Arabidopsis thaliana ubiquitin-ligase with components of the Saccharomyces cerevisiae ubiquitination machinery. Curr Genet 50:257-268. Altmann, R. 1890. Die Elementarorganisem und ihre Beziehungen zu den Zellen. Leipzig: Veit. Amerik, A., N. Sindhi, and M. Hochstrasser. 2006. A conserved late endosometargeting signal required for Doa4 deubiquitylating enzyme function. J Cell Biol 175:825-835. Amerik, A., S. Swaminathan, B. A. Krantz, K. D. Wilkinson, and M. Hochstrasser. 1997. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. Embo J 16:4826-4838. Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem 381:981-992. Amerik, A. Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell 11:3365-3380. Arlt, H., R. Tauer, H. Feldmann, W. Neupert, and T. Langer. 1996. The YTA1012 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875-885. Athenstaedt, K., and G. Daum. 2006. The life cycle of neutral lipids: synthesis, storage and degradation. Cell Mol Life Sci 63:1355-1369. Baek, K. H., M. A. Mondoux, R. Jaster, E. Fire-Levin, and A. D. D'Andrea. 2001. DUB-2A, a new member of the DUB subfamily of hematopoietic deubiquitinating enzymes. Blood 98:636-642. Baerends, R. J., S. W. Rasmussen, R. E. Hilbrands, M. van der Heide, K. N. Faber, P. T. Reuvekamp, J. A. Kiel, J. M. Cregg, I. J. van der Klei, and M. Veenhuis. 1996. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem 271:8887-8894. Baker, A., and I. A. Sparkes. 2005. Peroxisome protein import: some answers, more questions. Curr Opin Plant Biol 8:640-647. Baker, R. T., J. W. Tobias, and A. Varshavsky. 1992. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem 267:23364-23375. Bartz, R., W. H. Li, B. Venables, J. K. Zehmer, M. R. Roth, R. Welti, R. G. Anderson, P. Liu, and K. D. Chapman. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res 48:837-847. Bascom, R. A., H. Chan, and R. A. Rachubinski. 2003. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants. Mol Biol Cell 14:939-957. Beller, M., K. Thiel, P. J. Thul, and H. Jackle. 2010. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett 584:2176-2182. Berndt, C., D. Bech-Otschir, W. Dubiel, and M. Seeger. 2002. Ubiquitin system: JAMMing in the name of the lid. Curr Biol 12:R815-817. Biardi, L., and S. K. Krisans. 1996. Compartmentalization of cholesterol biosynthesis. Conversion of mevalonate to farnesyl diphosphate occurs in the peroxisomes. J Biol Chem 271:1784-1788. CHAPTER 4. REFERENCES________________________________________________ 91 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. 34. 35. Binns, D., T. Januszewski, Y. Chen, J. Hill, V. S. Markin, Y. Zhao, C. Gilpin, K. D. Chapman, R. G. Anderson, and J. M. Goodman. 2006. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173:719-731. Birschmann, I., K. Rosenkranz, R. Erdmann, and W. H. Kunau. 2005. Structural and functional analysis of the interaction of the AAA-peroxins Pex1p and Pex6p. Febs J 272:47-58. Birschmann, I., A. K. Stroobants, M. van den Berg, A. Schafer, K. Rosenkranz, W. H. Kunau, and H. F. Tabak. 2003. Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol Biol Cell 14:2226-2236. Blanchette-Mackie, E. J., N. K. Dwyer, T. Barber, R. A. Coxey, T. Takeda, C. M. Rondinone, J. L. Theodorakis, A. S. Greenberg, and C. Londos. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res 36:1211-1226. Bohm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res 25:2464-2469. Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J 20:51875196. Borst, P. 1989. Peroxisome biogenesis revisited. Biochim Biophys Acta 1008:1-13. Brown, L. A., and A. Baker. 2003. Peroxisome biogenesis and the role of protein import. J Cell Mol Med 7:388-400. Brown, L. A., and A. Baker. 2008. Shuttles and cycles: transport of proteins into the peroxisome matrix (review). Mol Membr Biol 25:363-375. Carvalho, A. F., M. P. Pinto, C. P. Grou, I. S. Alencastre, M. Fransen, C. SaMiranda, and J. E. Azevedo. 2007. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem 282:31267-31272. Chapman, K. D., and R. N. Trelease. 1991. Acquisition of membrane lipids by differentiating glyoxysomes: role of lipid bodies. J Cell Biol 115:995-1007. Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. Cohen, M., F. Stutz, N. Belgareh, R. Haguenauer-Tsapis, and C. Dargemont. 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5:661-667. Cohen, M., F. Stutz, and C. Dargemont. 2003. Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J Biol Chem 278:51989-51992. Czabany, T., K. Athenstaedt, and G. Daum. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim Biophys Acta 1771:299-309. De Duve, C., and P. Baudhuin. 1966. Peroxisomes (microbodies and related particles). Physiol Rev 46:323-357. Debelyy, M. O., H. W. Platta, D. Saffian, A. Hensel, S. Thoms, H. E. Meyer, B. Warscheid, W. Girzalsky, and R. Erdmann. 2011. Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery. Journal of Biological Chemistry. Debelyy, M. O., S. Thoms, M. Connerth, G. Daum, and R. Erdmann. 2011. Involvement of the Saccharomyces cerevisiae Hydrolase Ldh1p in Lipid Homeostasis. Eukaryot Cell 10:776-781. CHAPTER 4. REFERENCES________________________________________________ 92 36. 37. 38. 39. 40. 41. 42. 43. 44. 45. 46. 47. 48. 49. 50. 51. 52. 53. Delker, C., B. K. Zolman, O. Miersch, and C. Wasternack. 2007. Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal beta-oxidation enzymes-additional proof by properties of pex6 and aim1. Phytochemistry 68:1642-1650. Digel, M., R. Ehehalt, and J. Fullekrug. 2010. Lipid droplets lighting up: insights from live microscopy. FEBS Lett 584:2168-2175. Dodt, G., and S. J. Gould. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135:1763-1774. Doss-Pepe, E. W., E. S. Stenroos, W. G. Johnson, and K. Madura. 2003. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol 23:6469-6483. Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. Eitzen, G. A., R. K. Szilard, and R. A. Rachubinski. 1997. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol 137:12651278. Elgersma, Y., L. Kwast, M. van den Berg, W. B. Snyder, B. Distel, S. Subramani, and H. F. Tabak. 1997. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S.cerevisiae, causes proliferation of the endoplasmic reticulum membrane. Embo J 16:7326-7341. Erdmann, R., and G. Blobel. 1996. Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol 135:111-121. Erdmann, R., and W. Schliebs. 2005. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol 6:738-742. Ernst, R., B. Mueller, H. L. Ploegh, and C. Schlieker. 2009. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell 36:28-38. Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 35:93-114. Faber, K. N., J. A. Heyman, and S. Subramani. 1998. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol 18:936-943. Fan, W., and Y. Fujiki. 2006. A temperature-sensitive CHO pex1 mutant with a novel mutation in the AAA Walker A1 motif. Biochem Biophys Res Commun 345:1434-1439. Fang, Y., J. C. Morrell, J. M. Jones, and S. J. Gould. 2004. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol 164:863-875. Farese, R. V., Jr., and T. C. Walther. 2009. Lipid droplets finally get a little R-E-SP-E-C-T. Cell 139:855-860. Ferre, P. 2004. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes 53 Suppl 1:S4350. Fransen, M., I. Vastiau, C. Brees, V. Brys, G. P. Mannaerts, and P. P. Van Veldhoven. 2004. Potential role for Pex19p in assembly of PTS-receptor docking complexes. J Biol Chem 279:12615-12624. Fransen, M., I. Vastiau, C. Brees, V. Brys, G. P. Mannaerts, and P. P. Van Veldhoven. 2005. Analysis of human Pex19p's domain structure by pentapeptide scanning mutagenesis. J Mol Biol 346:1275-1286. CHAPTER 4. REFERENCES________________________________________________ 93 54. 55. 56. 57. 58. 59. 60. 61. 62. 63. 64. 65. 66. 67. 68. 69. 70. 71. 72. Fransen, M., T. Wylin, C. Brees, G. P. Mannaerts, and P. P. Van Veldhoven. 2001. Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol Cell Biol 21:4413-4424. Fujimoto, T., Y. Ohsaki, J. Cheng, M. Suzuki, and Y. Shinohara. 2008. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol 130:263-279. Funato, M., N. Shimozawa, T. Nagase, Y. Takemoto, Y. Suzuki, Y. Imamura, T. Matsumoto, T. Tsukamoto, T. Kojidani, T. Osumi, T. Fukao, and N. Kondo. 2006. Aberrant peroxisome morphology in peroxisomal beta-oxidation enzyme deficiencies. Brain Dev 28:287-292. Furuki, S., S. Tamura, N. Matsumoto, N. Miyata, A. Moser, H. W. Moser, and Y. Fujiki. 2006. Mutations in the peroxin Pex26p responsible for peroxisome biogenesis disorders of complementation group 8 impair its stability, peroxisomal localization, and interaction with the Pex1p x Pex6p complex. J Biol Chem 281:1317-1323. Gabaldon, T., B. Snel, F. van Zimmeren, W. Hemrika, H. Tabak, and M. A. Huynen. 2006. Origin and evolution of the peroxisomal proteome. Biol Direct 1:8. Gardocki, M. E., M. Bakewell, D. Kamath, K. Robinson, K. Borovicka, and J. M. Lopes. 2005. Genomic analysis of PIS1 gene expression. Eukaryot Cell 4:604-614. Ghaedi, K., S. Tamura, K. Okumoto, Y. Matsuzono, and Y. Fujiki. 2000. The peroxin pex3p initiates membrane assembly in peroxisome biogenesis. Mol Biol Cell 11:2085-2102. Gilchrist, C. A., and R. T. Baker. 2000. Characterization of the ubiquitin-specific protease activity of the mouse/human Unp/Unph oncoprotein. Biochim Biophys Acta 1481:297-309. Glickman, M. H., D. M. Rubin, V. A. Fried, and D. Finley. 1998. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol 18:3149-3162. Goldberg, I. J., and M. Merkel. 2001. Lipoprotein lipase: physiology, biochemistry, and molecular biology. Front Biosci 6:D388-405. Goodman, J. M. 2008. The gregarious lipid droplet. J Biol Chem 283:28005-28009. Goodman, J. M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res 50:2148-2156. Gorgas, K. 1986. Morphogenesis of peroxisomes in lipid synthesizing epithelia. Peroxisomes in Biology and Medicine H. D. Fahimi and H. Sies, editors. SpringerVerlag, Berlin, Heidelberg.:3-17. Gotte, K., W. Girzalsky, M. Linkert, E. Baumgart, S. Kammerer, W. H. Kunau, and R. Erdmann. 1998. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol 18:616-628. Gould, S. J., J. E. Kalish, J. C. Morrell, J. Bjorkman, A. J. Urquhart, and D. I. Crane. 1996. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTs1 receptor. J Cell Biol 135:85-95. Gouveia, A. M., C. P. Guimaraes, M. E. Oliveira, C. Reguenga, C. Sa-Miranda, and J. E. Azevedo. 2003. Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J Biol Chem 278:226-232. Gray, W. M., and J. S. Fassler. 1996. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol Cell Biol 16:347-358. Grou, C. P., A. F. Carvalho, M. P. Pinto, S. J. Huybrechts, C. Sa-Miranda, M. Fransen, and J. E. Azevedo. 2009. Properties of the ubiquitin-pex5p thiol ester conjugate. J Biol Chem 284:10504-10513. Grunau, S., W. Schliebs, R. Linnepe, C. Neufeld, C. Cizmowski, B. Reinartz, H. E. Meyer, B. Warscheid, W. Girzalsky, and R. Erdmann. 2009. Peroxisomal targeting of PTS2 pre-import complexes in the yeast Saccharomyces cerevisiae. Traffic 10:451-460. CHAPTER 4. REFERENCES________________________________________________ 94 73. 74. 75. 76. 77. 78. 79. 80. 81. 82. 83. 84. 85. 86. 87. 88. 89. 90. Guo, Y., K. R. Cordes, R. V. Farese, Jr., and T. C. Walther. 2009. Lipid droplets at a glance. J Cell Sci 122:749-752. Guterman, A., and M. H. Glickman. 2004. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem 279:17291738. Haan, G. J., K. N. Faber, R. J. Baerends, A. Koek, A. Krikken, J. A. Kiel, I. J. van der Klei, and M. Veenhuis. 2002. Hansenula polymorpha Pex3p is a peripheral component of the peroxisomal membrane. J Biol Chem 277:26609-26617. Hajra, A. K., and J. E. Bishop. 1982. Glycerolipid biosynthesis in peroxisomes via the acyl dihydroxyacetone phosphate pathway. Ann N Y Acad Sci 386:170-182. Hansen-Hagge, T. E., J. W. Janssen, H. Hameister, F. R. Papa, U. Zechner, T. Seriu, A. Jauch, D. Becke, M. Hochstrasser, and C. R. Bartram. 1998. An evolutionarily conserved gene on human chromosome 5q33-q34, UBH1, encodes a novel deubiquitinating enzyme. Genomics 49:411-418. Hayashi, Y., M. Hayashi, H. Hayashi, I. Hara-Nishimura, and M. Nishimura. 2001. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218:83-94. Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGAassociated Ubp8. Genes Dev 17:2648-2663. Heupel, R., and H. W. Heldt. 1994. Protein organization in the matrix of leaf peroxisomes. A multi-enzyme complex involved in photorespiratory metabolism. Eur J Biochem 220:165-172. Hicke, L. 2001. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2:195201. Hiraga, K., K. Suzuki, E. Tsuchiya, and T. Miyakawa. 1993. Cloning and characterization of the elongation factor EF-1 beta homologue of Saccharomyces cerevisiae. EF-1 beta is essential for growth. FEBS Lett 316:165-169. Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu Rev Genet 30:405-439. Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H. F. Tabak. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122:85-95. Hofmann, L., R. Saunier, R. Cossard, M. Esposito, T. Rinaldi, and A. Delahodde. 2009. A nonproteolytic proteasome activity controls organelle fission in yeast. J Cell Sci 122:3673-3683. Hohfeld, J., M. Veenhuis, and W. H. Kunau. 1991. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol 114:1167-1178. Honsho, M., T. Hiroshige, and Y. Fujiki. 2002. The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J Biol Chem 277:44513-44524. Honsho, M., S. Tamura, N. Shimozawa, Y. Suzuki, N. Kondo, and Y. Fujiki. 1998. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet 63:1622-1630. Hsiang, Y. H., R. Hertzberg, S. Hecht, and L. F. Liu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260:14873-14878. Hu, M., P. Li, M. Li, W. Li, T. Yao, J. W. Wu, W. Gu, R. E. Cohen, and Y. Shi. 2002. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041-1054. CHAPTER 4. REFERENCES________________________________________________ 95 91. 92. 93. 94. 95. 96. 97. 98. 99. 100. 101. 102. 103. 104. 105. 106. 107. Huang, Y., R. T. Baker, and J. A. Fischer-Vize. 1995. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science 270:1828-1831. Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. Hunt, J. E., and R. N. Trelease. 2004. Sorting pathway and molecular targeting signals for the Arabidopsis peroxin 3. Biochem Biophys Res Commun 314:586-596. Imanishi, Y., V. Gerke, and K. Palczewski. 2004. Retinosomes: new insights into intracellular managing of hydrophobic substances in lipid bodies. J Cell Biol 166:447453. Ingelmo-Torres, M., E. Gonzalez-Moreno, A. Kassan, M. Hanzal-Bayer, F. Tebar, A. Herms, T. Grewal, J. F. Hancock, C. Enrich, M. Bosch, S. P. Gross, R. G. Parton, and A. Pol. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic 10:1785-1801. Islinger, M., M. J. Cardoso, and M. Schrader. 2010. Be different--the diversity of peroxisomes in the animal kingdom. Biochim Biophys Acta 1803:881-897. Jakobs, B. S., and R. J. Wanders. 1991. Conclusive evidence that very-long-chain fatty acids are oxidized exclusively in peroxisomes in human skin fibroblasts. Biochem Biophys Res Commun 178:842-847. Jentsch, S., and S. Rumpf. 2007. Cdc48 (p97): a "molecular gearbox" in the ubiquitin pathway? Trends Biochem Sci 32:6-11. Johnson, M. A., W. B. Snyder, J. L. Cereghino, M. Veenhuis, S. Subramani, and J. M. Cregg. 2001. Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast 18:621-641. Jones, J. M., J. C. Morrell, and S. J. Gould. 2004. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol 164:57-67. Kahana, A. 2001. The deubiquitinating enzyme Dot4p is involved in regulating nutrient uptake. Biochem Biophys Res Commun 282:916-920. Kahana, A., and D. E. Gottschling. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol 19:6608-6620. Kal, A. J., A. J. van Zonneveld, V. Benes, M. van den Berg, M. G. Koerkamp, K. Albermann, N. Strack, J. M. Ruijter, A. Richter, B. Dujon, W. Ansorge, and H. F. Tabak. 1999. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell 10:1859-1872. Kammerer, S., N. Arnold, W. Gutensohn, H. W. Mewes, W. H. Kunau, G. Hofler, A. A. Roscher, and A. Braun. 1997. Genomic organization and molecular characterization of a gene encoding HsPXF, a human peroxisomal farnesylated protein. Genomics 45:200-210. Kashiwayama, Y., K. Asahina, H. Shibata, M. Morita, A. C. Muntau, A. A. Roscher, R. J. Wanders, N. Shimozawa, M. Sakaguchi, H. Kato, and T. Imanaka. 2005. Role of Pex19p in the targeting of PMP70 to peroxisome. Biochim Biophys Acta 1746:116-128. Kiel, J. A., K. Emmrich, H. E. Meyer, and W. H. Kunau. 2005. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem 280:1921-1930. Kiel, J. A., M. Veenhuis, and I. J. van der Klei. 2006. PEX genes in fungal genomes: common, rare or redundant. Traffic 7:1291-1303. CHAPTER 4. REFERENCES________________________________________________ 96 108. 109. 110. 111. 112. 113. 114. 115. 116. 117. 118. 119. 120. 121. 122. Kim, P. K., R. T. Mullen, U. Schumann, and J. Lippincott-Schwartz. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16dependent pathway from the ER. J Cell Biol 173:521-532. Kinner, A., and R. Kolling. 2003. The yeast deubiquitinating enzyme Ubp16 is anchored to the outer mitochondrial membrane. FEBS Lett 549:135-140. Knab, A. M., J. Fertala, and M. A. Bjornsti. 1993. Mechanisms of camptothecin resistance in yeast DNA topoisomerase I mutants. J Biol Chem 268:22322-22330. Knab, A. M., J. Fertala, and M. A. Bjornsti. 1995. A camptothecin-resistant DNA topoisomerase I mutant exhibits altered sensitivities to other DNA topoisomerase poisons. J Biol Chem 270:6141-6148. Kohlwein, S. D. 2010. Triacylglycerol homeostasis: insights from yeast. J Biol Chem 285:15663-15667. Koller, A., W. B. Snyder, K. N. Faber, T. J. Wenzel, L. Rangell, G. A. Keller, and S. Subramani. 1999. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol 146:99-112. Komori, M., J. A. Kiel, and M. Veenhuis. 1999. The peroxisomal membrane protein Pex14p of Hansenula polymorpha is phosphorylated in vivo. FEBS Lett 457:397-399. Kouranti, I., J. R. McLean, A. Feoktistova, P. Liang, A. E. Johnson, R. H. Roberts-Galbraith, and K. L. Gould. 2010. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol 8. Kragt, A., T. Voorn-Brouwer, M. van den Berg, and B. Distel. 2005. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem 280:7867-7874. Krahmer, N., Y. Guo, R. V. Farese, Jr., and T. C. Walther. 2009. SnapShot: Lipid Droplets. Cell 139:1024-1024 e1021. Krisans, S. K. 1992. The role of peroxisomes in cholesterol metabolism. Am J Respir Cell Mol Biol 7:358-364. Krisans, S. K. 1996. Cell compartmentalization of cholesterol biosynthesis. Ann N Y Acad Sci 804:142-164. Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. Lageweg, W., J. E. Sykes, M. Lopes-Cardozo, and R. J. Wanders. 1991. Oxidation of very-long-chain fatty acids in rat brain: cerotic acid is beta-oxidized exclusively in rat brain peroxisomes. Biochim Biophys Acta 1085:381-384. Lam, M. H., D. Urban-Grimal, A. Bugnicourt, J. F. Greenblatt, R. HaguenauerTsapis, and A. Emili. 2009. Interaction of the deubiquitinating enzyme Ubp2 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One 4:e4259. CHAPTER 4. REFERENCES________________________________________________ 97 123. 124. 125. 126. 127. 128. 129. 130. 131. 132. 133. 134. 135. 136. 137. 138. Lam, S. K., N. Yoda, and R. Schekman. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci U S A 107:21523-21528. Lametschwandtner, G., C. Brocard, M. Fransen, P. Van Veldhoven, J. Berger, and A. Hartig. 1998. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem 273:33635-33643. Layfield, R., K. Franklin, M. Landon, G. Walker, P. Wang, R. Ramage, A. Brown, S. Love, K. Urquhart, T. Muir, R. Baker, and R. J. Mayer. 1999. Chemically synthesized ubiquitin extension proteins detect distinct catalytic capacities of deubiquitinating enzymes. Anal Biochem 274:40-49. Lazarow, P. B., and W. H. Kunau. 1997. Peroxisomes. In E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.:547-605. Legakis, J. E., J. I. Koepke, C. Jedeszko, F. Barlaskar, L. J. Terlecky, H. J. Edwards, P. A. Walton, and S. R. Terlecky. 2002. Peroxisome senescence in human fibroblasts. Mol Biol Cell 13:4243-4255. Leon, S., L. Zhang, W. H. McDonald, J. Yates, 3rd, J. M. Cregg, and S. Subramani. 2006. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitindependent localization and regulation. J Cell Biol 172:67-78. Lin, Y., L. Sun, L. V. Nguyen, R. A. Rachubinski, and H. M. Goodman. 1999. The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284:328-330. Lindsey, D. F., A. Amerik, W. J. Deery, J. D. Bishop, M. Hochstrasser, and R. H. Gomer. 1998. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in Dictyostelium. J Biol Chem 273:2917829187. Linghu, B., J. Callis, and M. G. Goebl. 2002. Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryot Cell 1:491494. Listenberger, L. L., and D. A. Brown. 2008. Lipid droplets. Curr Biol 18:R237-238. Liu, Y., F. Wang, H. Zhang, H. He, L. Ma, and X. W. Deng. 2008. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J 55:844856. Lopez, R., V. Silventoinen, S. Robinson, A. Kibria, and W. Gish. 2003. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res 31:3795-3798. Lucau-Danila, A., T. Delaveau, G. Lelandais, F. Devaux, and C. Jacq. 2003. Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J Biol Chem 278:52641-52650. Ma, C., and S. Subramani. 2009. Peroxisome matrix and membrane protein biogenesis. IUBMB Life 61:713-722. Makarova, K. S., L. Aravind, and E. V. Koonin. 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci 25:50-52. Managadze, D., C. Wurtz, S. Wiese, M. Schneider, W. Girzalsky, H. E. Meyer, R. Erdmann, B. Warscheid, and H. Rottensteiner. 2010. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol 89:955-964. CHAPTER 4. REFERENCES________________________________________________ 98 139. 140. 141. 142. 143. 144. 145. 146. 147. 148. 149. 150. 151. 152. 153. 154. Mancuso, D. J., X. Han, C. M. Jenkins, J. J. Lehman, N. Sambandam, H. F. Sims, J. Yang, W. Yan, K. Yang, K. Green, D. R. Abendschein, J. E. Saffitz, and R. W. Gross. 2007. Dramatic accumulation of triglycerides and precipitation of cardiac hemodynamic dysfunction during brief caloric restriction in transgenic myocardium expressing human calcium-independent phospholipase A2gamma. J Biol Chem 282:9216-9227. Mancuso, D. J., C. M. Jenkins, H. F. Sims, J. M. Cohen, J. Yang, and R. W. Gross. 2004. Complex transcriptional and translational regulation of iPLAgamma resulting in multiple gene products containing dual competing sites for mitochondrial or peroxisomal localization. Eur J Biochem 271:4709-4724. Mancuso, D. J., H. F. Sims, X. Han, C. M. Jenkins, S. P. Guan, K. Yang, S. H. Moon, T. Pietka, N. A. Abumrad, P. H. Schlesinger, and R. W. Gross. 2007. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J Biol Chem 282:34611-34622. Marston, A. L., W. H. Tham, H. Shah, and A. Amon. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367-1370. Martin, S., and R. G. Parton. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7:373-378. Marzioch, M., R. Erdmann, M. Veenhuis, and W. H. Kunau. 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacylCoA thiolase, a PTS2-containing protein, into peroxisomes. Embo J 13:4908-4918. Matsumoto, N., S. Tamura, and Y. Fujiki. 2003. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 5:454-460. Matsuzono, Y., N. Kinoshita, S. Tamura, N. Shimozawa, M. Hamasaki, K. Ghaedi, R. J. Wanders, Y. Suzuki, N. Kondo, and Y. Fujiki. 1999. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci U S A 96:2116-2121. Matsuzono, Y., T. Matsuzaki, and Y. Fujiki. 2006. Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J Cell Sci 119:3539-3550. McCullock, S., T. Kinard, L. McCullough, and T. Formosa. 2006. blm3-1 is an allele of UBP3, a ubiquitin protease that appears to act during transcription of damaged DNA. J Mol Biol 363:660-672. McDonald, H. B., and B. Byers. 1997. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol 137:539-553. Miyata, N., and Y. Fujiki. 2005. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol 25:10822-10832. Motley, A., E. Hettema, B. Distel, and H. Tabak. 1994. Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol 125:755-767. Motley, A. M., and E. H. Hettema. 2007. Yeast peroxisomes multiply by growth and division. J Cell Biol 178:399-410. Muller, W. H., T. P. van der Krift, A. J. Krouwer, H. A. Wosten, L. H. van der Voort, E. B. Smaal, and A. J. Verkleij. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. Embo J 10:489-495. Mullner, H., D. Zweytick, R. Leber, F. Turnowsky, and G. Daum. 2004. Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1663:9-13. CHAPTER 4. REFERENCES________________________________________________ 99 155. 156. 157. 158. 159. 160. 161. 162. 163. 164. 165. 166. 167. 168. 169. 170. 171. 172. 173. Murakami, M., S. Masuda, K. Ueda-Semmyo, E. Yoda, H. Kuwata, Y. Takanezawa, J. Aoki, H. Arai, H. Sumimoto, Y. Ishikawa, T. Ishii, Y. Nakatani, and I. Kudo. 2005. Group VIB Ca2+-independent phospholipase A2gamma promotes cellular membrane hydrolysis and prostaglandin production in a manner distinct from other intracellular phospholipases A2. J Biol Chem 280:14028-14041. Neuberger, G., S. Maurer-Stroh, B. Eisenhaber, A. Hartig, and F. Eisenhaber. 2003. Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxonspecific differences. J Mol Biol 328:567-579. Neuberger, G., S. Maurer-Stroh, B. Eisenhaber, A. Hartig, and F. Eisenhaber. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J Mol Biol 328:581-592. Niederhoff, K., N. M. Meindl-Beinker, D. Kerssen, U. Perband, A. Schafer, W. Schliebs, and W. H. Kunau. 2005. Yeast Pex14p possesses two functionally distinct Pex5p and one Pex7p binding sites. J Biol Chem 280:35571-35578. Novikoff, A. B., P. M. Novikoff, O. M. Rosen, and C. S. Rubin. 1980. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol 87:180-196. Nyathi, Y., and A. Baker. 2006. Plant peroxisomes as a source of signalling molecules. Biochim Biophys Acta 1763:1478-1495. Papa, F. R., A. Y. Amerik, and M. Hochstrasser. 1999. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell 10:741-756. Papa, F. R., and M. Hochstrasser. 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366:313-319. Park, S., R. Isaacson, H. T. Kim, P. A. Silver, and G. Wagner. 2005. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13:9951005. Pernas, M. A., C. Lopez, L. Pastrana, and M. L. Rua. 2001. Purification and characterization of Lip2 and Lip3 isoenzymes from a Candida rugosa pilot-plant scale fed-batch fermentation. J Biotechnol 84:163-174. Petriv, O. I., and R. A. Rachubinski. 2004. Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. J Biol Chem 279:19996-20001. Petriv, O. I., L. Tang, V. I. Titorenko, and R. A. Rachubinski. 2004. A new definition for the consensus sequence of the peroxisome targeting signal type 2. J Mol Biol 341:119-134. Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503-533. Platta, H. W., M. O. Debelyy, F. El Magraoui, and R. Erdmann. 2008. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans 36:99-104. Platta, H. W., F. El Magraoui, D. Schlee, S. Grunau, W. Girzalsky, and R. Erdmann. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol 177:197-204. Platta, H. W., and R. Erdmann. 2007. The peroxisomal protein import machinery. FEBS Lett 581:2811-2819. Platta, H. W., W. Girzalsky, and R. Erdmann. 2004. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J 384:37-45. Platta, H. W., S. Grunau, K. Rosenkranz, W. Girzalsky, and R. Erdmann. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol 7:817-822. Ploegh, H. L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448:435-438. CHAPTER 4. REFERENCES________________________________________________ 100 174. 175. 176. 177. 178. 179. 180. 181. 182. 183. 184. 185. 186. 187. 188. 189. 190. 191. 192. Purdue, P. E., and P. B. Lazarow. 2001. Pex18p is constitutively degraded during peroxisome biogenesis. J Biol Chem 276:47684-47689. Rajakumari, S., K. Grillitsch, and G. Daum. 2008. Synthesis and turnover of nonpolar lipids in yeast. Prog Lipid Res 47:157-171. Rayapuram, N., and S. Subramani. 2006. The importomer--a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim Biophys Acta 1763:1613-1619. Rhodin, J. 1954. Correlation of ultrastructural organization and function in normal and experimentally changed proximal convoluted tubule cells of the mouse kidney. Aktiebolaget Godvil, Stockholm. Rosenkranz, K., I. Birschmann, S. Grunau, W. Girzalsky, W. H. Kunau, and R. Erdmann. 2006. Functional association of the AAA complex and the peroxisomal importomer. Febs J 273:3804-3815. Rucktaschel, R., W. Girzalsky, and R. Erdmann. 2011. Protein import machineries of peroxisomes. Biochim Biophys Acta 1808:892-900. Rucktaschel, R., S. Thoms, V. Sidorovitch, A. Halbach, M. Pechlivanis, R. Volkmer, K. Alexandrov, J. Kuhlmann, H. Rottensteiner, and R. Erdmann. 2009. Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis. J Biol Chem 284:20885-20896. Rumpf, S., and S. Jentsch. 2006. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21:261-269. Sakamoto, T., T. Tanaka, Y. Ito, S. Rajesh, M. Iwamoto-Sugai, Y. Kodera, N. Tsuchida, T. Shibata, and T. Kohno. 1999. An NMR analysis of ubiquitin recognition by yeast ubiquitin hydrolase: evidence for novel substrate recognition by a cysteine protease. Biochemistry 38:11634-11642. Santos, M. J., T. Imanaka, H. Shio, G. M. Small, and P. B. Lazarow. 1988. Peroxisomal membrane ghosts in Zellweger syndrome--aberrant organelle assembly. Science 239:1536-1538. Schliebs, W., W. Girzalsky, and R. Erdmann. 2010. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol 11:885-890. Schliebs, W., and W. H. Kunau. 2004. Peroxisome membrane biogenesis: the stage is set. Curr Biol 14:R397-399. Schluter, A., S. Fourcade, R. Ripp, J. L. Mandel, O. Poch, and A. Pujol. 2006. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol 23:838-845. Schrader, M. 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J Histochem Cytochem 49:1421-1429. Schrader, M., and H. D. Fahimi. 2008. The peroxisome: still a mysterious organelle. Histochem Cell Biol 129:421-440. Schrag, J. D., and M. Cygler. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol 284:85-107. Schueller, N., S. J. Holton, K. Fodor, M. Milewski, P. Konarev, W. A. Stanley, J. Wolf, R. Erdmann, W. Schliebs, Y. H. Song, and M. Wilmanns. 2010. The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. Embo J 29:2491-2500. Schulz, H. 1991. Beta oxidation of fatty acids. Biochim Biophys Acta 1081:109-120. Shibata, H., Y. Kashiwayama, T. Imanaka, and H. Kato. 2004. Domain architecture and activity of human Pex19p, a chaperone-like protein for intracellular trafficking of peroxisomal membrane proteins. J Biol Chem 279:38486-38494. CHAPTER 4. REFERENCES________________________________________________ 101 193. 194. 195. 196. 197. 198. 199. 200. 201. 202. 203. 204. 205. 206. 207. 208. 209. Shiozawa, K., N. Maita, K. Tomii, A. Seto, N. Goda, Y. Akiyama, T. Shimizu, M. Shirakawa, and H. Hiroaki. 2004. Structure of the N-terminal domain of PEX1 AAA-ATPase. Characterization of a putative adaptor-binding domain. J Biol Chem 279:50060-50068. Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. Skinner, J. R., T. M. Shew, D. M. Schwartz, A. Tzekov, C. M. Lepus, N. A. Abumrad, and N. E. Wolins. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem 284:30941-30948. Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol 158:259-271. Smith, J. J., S. A. Ramsey, M. Marelli, B. Marzolf, D. Hwang, R. A. Saleem, R. A. Rachubinski, and J. D. Aitchison. 2007. Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol Syst Biol 3:115. Snyder, W. B., A. Koller, A. J. Choy, and S. Subramani. 2000. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol 149:1171-1178. Soukupova, M., C. Sprenger, K. Gorgas, W. H. Kunau, and G. Dodt. 1999. Identification and characterization of the human peroxin PEX3. Eur J Cell Biol 78:357-374. South, S. T., and S. J. Gould. 1999. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol 144:255-266. Stobart, A. K., S. Stymne, and S. Höglund. 1986. Safflower microsomes catalyse oil accumulation in vitro: A model system. Planta 169:33-37. Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell 10:2583-2594. Tamura, S., N. Shimozawa, Y. Suzuki, T. Tsukamoto, T. Osumi, and Y. Fujiki. 1998. A cytoplasmic AAA family peroxin, Pex1p, interacts with Pex6p. Biochem Biophys Res Commun 245:883-886. Tamura, S., S. Yasutake, N. Matsumoto, and Y. Fujiki. 2006. Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J Biol Chem 281:27693-27704. Tauchi-Sato, K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem 277:44507-44512. Terlecky, S. R., J. I. Koepke, and P. A. Walton. 2006. Peroxisomes and aging. Biochim Biophys Acta 1763:1749-1754. Thiele, C., and J. Spandl. 2008. Cell biology of lipid droplets. Curr Opin Cell Biol 20:378-385. Thoms, S. 2002. Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett 520:107-110. Thoms, S., M. O. Debelyy, M. Connerth, G. Daum, and R. Erdmann. 2011. The Putative Saccharomyces cerevisiae Hydrolase Ldh1p Is Localized to Lipid Droplets. Eukaryot Cell 10:770-775. CHAPTER 4. REFERENCES________________________________________________ 102 210. 211. 212. 213. 214. 215. 216. 217. 218. 219. 220. 221. 222. 223. 224. 225. 226. 227. 228. 229. 230. 231. Thoms, S., M. O. Debelyy, K. Nau, H. E. Meyer, and R. Erdmann. 2008. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. Febs J 275:504514. Thoms, S., and R. Erdmann. 2005. Import of proteins into peroxisomes. Movement Across Membranes (Eichler J, ed.) Springer, New York.:125–134. Thoms, S., and R. Erdmann. 2006. Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta 1763:1620-1628. Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. Embo J 19:94-102. Tobias, J. W., and A. Varshavsky. 1991. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem 266:12021-12028. Tolbert, N. E. 1981. Metabolic pathways in peroxisomes and glyoxysomes. Annu Rev Biochem 50:133-157. Tolbert, N. E., and E. Essner. 1981. Microbodies: peroxisomes and glyoxysomes. J Cell Biol 91:271s-283s. Tower, R. J., A. Fagarasanu, J. D. Aitchison, and R. A. Rachubinski. 2011. The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol Biol Cell 22:1727-1738. van den Bosch, H., R. B. Schutgens, R. J. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu Rev Biochem 61:157-197. van Roermund, C. W., E. H. Hettema, A. J. Kal, M. van den Berg, H. F. Tabak, and R. J. Wanders. 1998. Peroxisomal beta-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. Embo J 17:677-687. Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates, 3rd, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. Walther, T. C., and R. V. Farese, Jr. 2009. The life of lipid droplets. Biochim Biophys Acta 1791:459-466. Walton, P. A., P. E. Hill, and S. Subramani. 1995. Import of stably folded proteins into peroxisomes. Mol Biol Cell 6:675-683. Wanders, R. J. 2004. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol Genet Metab 83:16-27. Wang, Q., L. Li, and Y. Ye. 2008. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem 283:7445-7454. Weber, H. 2002. Fatty acid-derived signals in plants. Trends Plant Sci 7:217-224. Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169-178. Welchman, R. L., C. Gordon, and R. J. Mayer. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6:599-609. Weller, S., I. Cajigas, J. Morrell, C. Obie, G. Steel, S. J. Gould, and D. Valle. 2005. Alternative splicing suggests extended function of PEX26 in peroxisome biogenesis. Am J Hum Genet 76:987-1007. Weller, S., S. J. Gould, and D. Valle. 2003. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet 4:165-211. Welte, M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol 17:363-369. Wiebel, F. F., and W. H. Kunau. 1992. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature 359:73-76. CHAPTER 4. REFERENCES________________________________________________ 103 232. 233. 234. 235. 236. 237. 238. 239. Williams, C., M. van den Berg, R. R. Sprenger, and B. Distel. 2007. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 282:22534-22543. Wilson, E. 1896. The Cell in Development and Inheritance. New York: Macmillan. Yang, J., X. Han, and R. W. Gross. 2003. Identification of hepatic peroxisomal phospholipase A(2) and characterization of arachidonic acid-containing choline glycerophospholipids in hepatic peroxisomes. FEBS Lett 546:247-250. Yao, T., and R. E. Cohen. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403-407. Ye, Y., H. H. Meyer, and T. A. Rapoport. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162:7184. Zehmer, J. K., R. Bartz, P. Liu, and R. G. Anderson. 2008. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci 121:1852-1860. Zhang, J. W., Y. Han, and P. B. Lazarow. 1993. Novel peroxisome clustering mutants and peroxisome biogenesis mutants of Saccharomyces cerevisiae. J Cell Biol 123:1133-1147. Zweytick, D., K. Athenstaedt, and G. Daum. 2000. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta 1469:101-120. CHAPTER 5. MISCELLANEOUS____________________________________________ 104 CHAPTER 5. MISCELLANEOUS 5.1 Publications 1. Debelyy, M. O., H. W. Platta, D. Saffian, A. Hensel, S. Thoms, H. E. Meyer, B. Warscheid, W. Girzalsky, and R. Erdmann. 2011. Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery. Journal of Biological Chemistry. In press 2. Debelyy, M. O., S. Thoms, M. Connerth, G. Daum, and R. Erdmann. 2011. Involvement of the Saccharomyces cerevisiae Hydrolase Ldh1p in Lipid Homeostasis. Eukaryot Cell 10:776-781. 3. Platta, H. W., M. O. Debelyy, F. El Magraoui, and R. Erdmann. 2008. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans 36:99-104. 4. Thoms, S., M. O. Debelyy, M. Connerth, G. Daum, and R. Erdmann. 2011. The Putative Saccharomyces cerevisiae Hydrolase Ldh1p Is Localized to Lipid Droplets. Eukaryot Cell 10:770-775. 5. Thoms, S., M. O. Debelyy, K. Nau, H. E. Meyer, and R. Erdmann. 2008. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. Febs J 275:504514. CHAPTER 5. MISCELLANEOUS____________________________________________ 105 5.2 Personal contribution to the papers 1. Debelyy, M. O., H. W. Platta, D. Saffian, A. Hensel, S. Thoms, H. E. Meyer, B. Warscheid, W. Girzalsky, and R. Erdmann. 2011. Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery. Journal of Biological Chemistry. In press. Planning: 70 % Experiments: 70 % Manuscript writing: 30 % 2. Debelyy, M. O., S. Thoms, M. Connerth, G. Daum, and R. Erdmann. 2011. Involvement of the Saccharomyces cerevisiae Hydrolase Ldh1p in Lipid Homeostasis. Eukaryot Cell 10:776-781. Planning: 80 % Experiments: 80 % Manuscript writing: 80 % 3. Platta, H. W., M. O. Debelyy, F. El Magraoui, and R. Erdmann. 2008. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans 36:99-104. Manuscript writing: 30 % 4. Thoms, S., M. O. Debelyy, M. Connerth, G. Daum, and R. Erdmann. 2011. The Putative Saccharomyces cerevisiae Hydrolase Ldh1p Is Localized to Lipid Droplets. Eukaryot Cell 10:770-775. Planning: 50 % Experiments: 50 % Manuscript writing: 20 % 5. Thoms, S., M. O. Debelyy, K. Nau, H. E. Meyer, and R. Erdmann. 2008. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. Febs J 275:504514. Planning: 50 % Experiments: 30 % Manuscript writing: 20 % CHAPTER 5. MISCELLANEOUS____________________________________________ 106 5.3 Conferences 1. Open European Peroxisome Meeting 2006. Leuven, Belgium, 18-19 September 2006. (Poster) 2. VAAM-Symposium: Biology of Yeast and Filamentous Fungi 2006. Bochum, Germany, 12 October 2006. (Poster) 3. Seventh International Meeting on AAA Proteins 2007. Royal Agricultural College, Cirencester, United Kingdom, 9—13 September 2007. (Poster) 4. The EMBO Meeting – Advancing the Life Sciences 2009. Amsterdam, Netherlands, 29 August – 1 September 2009. (Poster) CHAPTER 5. MISCELLANEOUS____________________________________________ 107 5.4 Curriculum Vitae PERSONAL DATA Name Mykhaylo O. Debelyy Date of Birth 11 March 1978 Place of Birth Dnepropetrovsk, Ukraine Citizenship Ukrainian Marital Status Married, one child EDUCATION July 2006 – July 2011 Ruhr-University Bochum Institute of Physiological Chemistry Department of System Biochemistry Ph.D. student Guidance by: Prof. Dr. Ralf Erdmann Dr. Wolfgang Girzalsky September 1996 – August 2001 Dnepropetrovsk National University Department of Biochemistry & Biophysics Dipl.-Biol. &. Biochem. Guidance by: Prof. Dr. Natalia I. Shtemenko Prof. Dr. Galina A. Ushakova CHAPTER 5. MISCELLANEOUS____________________________________________ 108 5.5 Acknowledgement Prof. Dr. Ralf Erdmann Vishal Kalel Alexander Neuhaus Prof. Dr. Wolf-H. Kunau Delia Saffian Prof. Dr. Günter Daum Immanuel Grimm Fouzi El Magraoui PD Dr. Mathias Lübben Sohel Hasan PD Dr. Wolfgang Schliebs Sabrina Mindthoff Sabrina Beck Dr. Wolfgang Girzalsky Rezeda Mirgalieva Imtiaz Ali Dr. Robert Rucktäschel Dr. Harald W. Platta Christiane Sprenger Dr. Shirisha Nagotu Sigrid Wuethrich Dr. Christian Cizmowski Monika Bürger Dr. Pratima Bharti Ülrike Freimann Dr. David Managadze Elisabeth Becker Frauke Albustin Meike Möller Britta Stickel CHAPTER 5. MISCELLANEOUS____________________________________________ 109 5.6 Global scientific outlook for human race Sociology: 1. Is it possible to create a classless society of equal possibilities for each human being, where all people have an access to high quality food, safe environment, accommodation, medical services, elementary and higher education and have the possibility for individual development? Biology: 1. What are life and death? 2. What is brain and what is the nature of consciousness? 3. Is it possible to improve the limited nature of human beings? Physics: 1. What is the time? 1. What is the nature of gravity and momentum? 2. What is the nature of electric and magnetic fields? 3. What is space and what it is filled with? Philosophy: 1. Is there a limit to the scientific exploration of the universe? If yes, then would it make sense to improve the limited nature of human beings by the physical influence?