* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Biochemistry
Isotopic labeling wikipedia , lookup
Gaseous signaling molecules wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Lipid signaling wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Biosequestration wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Paracrine signalling wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Microbial metabolism wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Phosphorylation wikipedia , lookup
Biochemical cascade wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Biosynthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Photosynthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
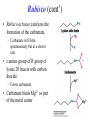
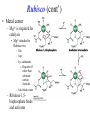
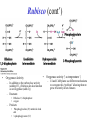
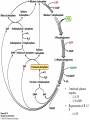
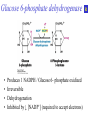
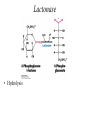
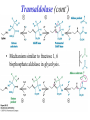
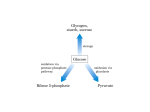
Biochemistry The Calvin Cycle and Pentose Phosphate Pathway Stryer Ch. 20 Calvin Cycle (Dark Reaction) • Reduce carbon dioxide to carbohydrates using NADPH and ATP produced during the light reactions of photosynthesis • Three Stages – Fix (attach) CO2 to Ribulose 1,5-bisphosphate forming two molecules of 3phosphoglycerate – Reduce 3-phosphoglycerate to form hexose carbohydrates – Regenerate ribulose 1,5bisphosphate Ribulose 1,5-bisphosphate carboxylase / oxygenase (Rubisco) • Structure – 8 large (L) subunits – 55 kd • 1 catalytic site / L subunit • 1 regulatory site / L subunit – 8 small (S) subunits – 13 kd • Most abundant enzyme on earth. – ~ 30% of leaf protein • Reaction rate is slow – Rate = 3 /s • ΔG °´= -51.9 kJ/mol (-12.4 kcal/mol) Rubisco (cont’) • Rubisco activase catalyzes the formation of the carbamate. – Carbamate will form spontaneously but at a slower rate. • ε amino group of R group of lysine 201reacts with carbon dioxide. – Forms carbamate • Carbamate binds Mg2+ as part of the metal center Rubisco (cont’) • Metal center – Mg2+ is required for catalysis • Mg2+ attached to Rubisco via: – Glu – Asp – Lys carbamate » Requires CO2 other than substrate carbon dioxides. – Also binds water – Ribulose 1,5bisphosphate binds and activates Rubisco (cont’) • Carboxylase Activity – Fix CO2 (1C) to Ribulose 1,5bisphosphate (5C) producing a unstable intermediate (6C) which reacts to form 2 molecules of 3phosphoglycerate. – Reactants – Ribulose 1,5-bisphosphate • In Stroma (25°C) • Carbon dioxide – [CO2]= 10 μM – Products – [O2]= 250 μM • 3-phosphoglycerate (2 – Favored over oxygenase activity molecules) Rubisco (cont’) • Oxygenase Activity – In addition to the carboxylase activity (adding CO2), Rubisco can also function as an oxygenase (adds O2) – Reactants • Ribulose 1,5-bisphosphate • oxygen – Products: • Phosphoglycolate (2C) metabolic dead end • 3-phosphoglycerate (3C) • Oxygenase activity ↑ as temperature ↑. – C4 and CAM plants use different mechanisms to overcome this “problem” allowing them to grow efficiently in hot climates. Photorespiratory Reactions • Recovers the carbon skeleton from phosphoglycolate • Chloroplast Phosphotase • Peroxisome (aka microsome) • Mitochondria Glycolate oxidase Reduction of 3phosphoglycerate to a hexose requires: • Reactants – 3-phosphoglycerate (2 molecules) • Product – Fructose 6-phosphate (1 molecule) inter-converts into other hexoses. • Energy provided by light reaction – ATP – NADPH Reduction of 3-phosphoglycerate to a hexose requires: • Transketolase – Transfers a COCH2OH (2C) unit from a ketose to an aldose. – TPP as coenzyme • Aldolase – (Do you remember ALDOLASE? An aldol condensation) – Transfers DHAP to an aldose of (n carbons) making a ketose (n +3C) • Specific for DHAP but accepts many aldehyde containing sugars Synthesis of C4, C5 and C7 sugars • The combined action of transketolase and aldolase as part of the Calvin cycle provide the plant with a source of C4, C5 and C7 sugars. Regenerate of ribulose 1,5 bisphosphate • Phosphopentose isomerase • Phosphopentose epimerase • Phosphoribulose kinase • 1 molecule glucose requires – 12 ATP – 12 NADPH • Regeneration of R 1,5 P – 6 ATP Synthesis of Starch • Polymer of glucose with alpha 1-4 glycosidic bonds and alpha 1-6 branches. – Fewer branches than glycogen – Activated precursor ADP-glucose Synthesis of Sucrose • synthesised in cytoplasm • Sucrose-6-phosphate synthase – fructose -6 phosphate • Synthesized from trioses (G3P) from ct – Phosphate translocator » Antiports phosphate / G3P – UDP-glucose Regulation of the Calvin Cycle • Light Reaction alters chemistry of stroma by: – ↑ [NADPH] • Reduction of NADP+ by FdNADP+ reductase • Activates two enzymes by displacing the inhibitory protein CP12 – Phosphoribulose kinase – Glyceraldehyde 3-phophate dehydrogenase – ↑ pH from 7 to 8 • Pumping H+ by cyt bf • Alkaline pH favors carbamate formation – ↑ [Mg2+] • Transported due to transport of H+ – ↑ reduced Fd • Reduces thioredoxin Thioredoxin: Structure • 12 kd protein • Pair of cysteine cycle between oxidized disulfides and reduced sulfhydryls • Contains 4Fe-4S cluster which couples two single e- reduction into a two ereduction. Regulation by Thioredoxin • Activates biosynthetic pathways by reducing regulatory enzymes of that pathway. • Ferridoxin reduces thioredoxin which reduces regulatory enzymes in pathway. The Location or Timing of the Calvin cycle relative to Carbon Fixation reduces Rubiso’s Oxagenase activity • (WHO) – C3, C4, CAM plants • (WHAT) – first molecule formed from CO2 • (WHEN) – molecule formed • (WHERE) – molecule formed Location/ Timing of Calvin cycle Differs in C3, C4, and CAM Plants • (WHY) – to increase the local [CO2] relative to [O2] • favors photosynthesis over photorespiration. • Atmosphere is: – 21% oxygen – 0.036% carbon dioxide C3 Plant (This is what we have been discussing) • Wheat, rice, oats, rose, soybeans • 3PG = 3 phosphoglycerate first molecule formed in Calvin Cycle RuBP Carboxylase RuBP + CO2 • Mesophyll cells – Carbon Dioxide fixation – Calvin cycle 3 PG C4 Plants • Corn, Sugar cane, Bermuda grass. • Oxaloacetate (C4) Phosphoenolpyruvate Carboxylase PEP (C3) + CO2 Oxaloacetate (C4) Pineapple CAM • Spatal (space/ location) separation CO2 Night Organic acid – mesophyll cells • carbon dioxide fixation – bundle sheath cells • Calvin cycle CO2 Day Calvin Cycle Sugar (b) Temporal separation of steps CAM (Crassulacean Acid Metabolism) • Succulent desert plants (cacti), pineapple • Oxaloacetate(C4) Malate (C4) Sugarcane C CO • Temporal (time) separation 4 2 Mesophyll cell Organic acid – Mesophyll cells 1 • night –Bundlestomata openCO-2 allows CO2 to enter sheath cell 2 – carbon fixation Calvin • day Cycle – stomata closed Sugar- conserves water – Calvin cycle (a) Spatial separation of steps Pentose Phosphate Pathway • AKA – Hexose Monophosphate Pathway – Phosphoglyconate Pathway – Pentose Monophosphate Shunt • Function: – Synthesis of NADPH • Reduction power for biosynthetic reactions. – Catabolism / synthesis of C 5 (pentoses) carbohydrates • Nucleotide biosynthesis – Catabolism / synthesis of C 4 (tetroses) and 7 C (heptoses) carbohydrates – Linked to glycolysis • Divided into – Oxidative phase • Synthesis of NADPH – Nonoxidative phase • Interconversion of C4, C5, C6, and C7 carbohydrates Pentose Phosphate Pathway • Location of Pentose phosphate pathway to provide NADPH: – Synthesis • • • • • • • • Adrenal gland - Steroid synthesis Testes - Steroid synthesis Ovary - Steroid synthesis Mammary gland - Fatty acid synthesis Liver - Fatty acid biosynthesis and Cholesterol biosynthesis Adipose tissue - Fatty acid synthesis Various tissue - Neurotransmitter biosynthesis Almost all tissue - Nucleotide biosynthesis – Detoxification • Reduction of oxidized glutathione • Cytochrome P 450 monooxygenase Enzymes of Pentose phosphate pathway Oxidative Phase Pentose Phosphate Pathway • Produces 1 Ribulose 5 – phosphate and reduces 2 NADPH / glucose 6-phosphate • Ribulose 5-phosphate (C5) produces – Ribose 5-phosphate (C5) – Xylulose 5-phosphate (C5) Glucose 6-phosphate dehydrogenase • • • • Produces 1 NADPH / Glucose 6- phosphate oxidized Irreversible Dehydrogenation Inhibited by ↓ [NADP+] (required to accept electrons) R Lactonase • Hydrolysis 6-Phosphogluconate dehydrogenase • Produces 1 NADPH / Glucose 6- phosphate oxidized Phosphopentose isomerase • Ribulose 5-phosphate → Ribose 5-phosphate Phosphopentose epimerase • Ribulose 5-phosphate → xylulose 5-phosphate Transketolase • Transfers a COCH2OH (2C) unit from a ketose to an aldose. • TPP as coenzyme • C5 + C5 ⇌ C3 + C7 Transketolase (cont’) • Mechanism similar to E1 subunit of pyruvate dehydrogenase complex. Transaldolase • Transfers 3C units • Uses R group of lysine to form Schiffs base (no prosthetic group) • C3 + C7 ⇌ C6 + C4 Transaldolase (cont’) • Mechanism similar to fructose 1, 6 bisphosphate aldolase in glycolysis. Transketolase • C4 + C5 ⇌ C6 + C3 Similarities in Transketolase and Transaldolase Mechanism • Both enzymes produce carbanions stabilized by resonance during catalysis. – Transketolase • TPP – Transaldolase • lysine Pentose Phosphate Pathway adjusts to needs of the cell • For production of NADPH or various carbohydrates Situation 1 High demand for ribose 5-phosphate (for DNA synthesis); low demand for NADPH • Glycolysis linked to transketolase and transaldolase – 2 molecules of fructose 6phosphate + 1 molecule of glyceraldehy de 3phosphate →3 molecules ribose 5phosphate. Situation 2 Balanced need for ribose 5-phosphate and NADPH. • Uses oxidative phase of pentose phosphate pathway to produce 2 NADPH and 1 ribose 5phosphate. Situation 3 More NADPH than ribose 5 phosphate required. • Glucose 6phosphate completely oxidized to CO2 • Three reactions – Oxidative phase of pentose phosphate pathway – Tranketolase and tranaldolase – Gluconeogenesis Situation 4 Both NADPH and ATP required • Oxidative phase of pentose phosphate pathway. • Ribose 5-phosphate converted into F6P and G3P which enter glycolysis • Pyruvate – Oxidized – Used as precursors Glutathione protects against Reactive oxygen species (ROS) • Glutathione – Tripeptide of ECG • Free sulfhydryl • GSH → GSSG – GSH = reduced glutathione – GSSG = oxidized glutathione • Glutathione reductase – Uses NADPH to reduce GSSG → GSH. – Contains FAD • In vivo – [glutathione] = ~ 5mM – [GSH] : [GSSG] 500 : 1 Glucose 6phosphate dehydrogenase deficiency • Glucose 6-phosphate dehydrogenase • X linked inheritance • MOST common genetic defect • Pamaquine – Purine glycoside isolated from a plant (fava bean) – Antimalarial drug – In vivo pamaquine generates ↑ [peroxides] – causes severe symptoms in some patients • Black urine • Jaundice • Hemolytic anemia • Symptoms due to glucose 6phosphate deficiency – Does not produce NADPH via pentose phosphate pathway – NADPH needed by glutathione reductase to reduce glutathione • Glutathione destroys ROS in RBCs • RBCs with Heinz bodies. – Defining characteristic of glucose 6phosphate dehydrogenase deficiency – Denatured proteins adhering to plasma membrane of RBC.