* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download in situ - Moodle NTOU
Long non-coding RNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Pathogenomics wikipedia , lookup
Copy-number variation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome evolution wikipedia , lookup
Public health genomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome (book) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
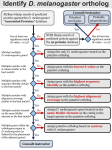
Bioinformatics Assignment • Project practice: – We are going to integrate the tools and idea you learned from this course, and apply them to finish a project. – In this project, you have to choose a human gene which you are interested. Then try to search the related genes from mouse and zebrafish. Try to dig out the resources from other animal models to enhance your understanding of your target gene. 1 Project Practice Report • The report should contain five parts followed by the instruction. • Each part will be count for 20% of your final score! • Please hand in you report as a print out hard copy to Prof. Tzou’s office before Jan 20, 2010. 2 Part 1: Find your target gene • What kind of human gene are you interested? – Gene Linked to disease? • Use OMIM to find your target! • Paste the OMIM description and translated it to Chinese in short! – Other reason • Please descript it! 3 OMIM • Search the human genetic diseases which you are interested! • http://www.ncbi.nlm.nih.gov/omim 4 Example of OMIM search result Gene Linked to this disease 5 Target Gene Product in FASTA Format • Select and copy protein sequence! 6 Part 2: Protein Domain Analysis • Use InterProScan to search important domain of your target protein! • Please Paste your search result in your report • Find another tool to do the same work. Also paste the result in your report. • Compare the results from the two different tools. 7 InterProScan • http://www.ebi.ac.uk/InterProScan / 8 InterProScan result 9 Part 3: Find out ortholog genes in mouse and zebrafish • You may use Panther classification system to find out related genes. • You may also use BLAST to search for related genes in mouse or zebrafish. • Please paste the MGI and Zfin Page of your target gene in your report. – Be careful to distinguish between ortholog and paralog genes 10 11 Panther Tree 12 ZFIN gene summary • http://zfin.org/cgi-bin/webdriver?MIval=aa-ZDB_home.apg 13 MGI gene summary • http://www.informatics.jax.org/ 14 Part 4: Resources from MGI and Zfin • Phenotype: – Knock out phenotype? – Knock down tool? – Please include these information in your report! • Expression Pattern: – – – – cDNA, RT-PCR, microarray and in situ hybridization EST profile in unigene Please include EST profiles of all ortholog genes. It is also welcome to put as much as expression information in your report. – You may compare the expression profile of ortholog genes to the human one and discuss about it! 15 Linked resource from MGI 16 Linked resources from ZFIN 17 UniGene 18 EST profile • You have to include pages of the EST profile from human, mouse and zebrafish ortholog genes! 19 Part 5: Open Question of Protein Analysis • Please try your best to dig out as many as possible information to compare the ortholog genes form mouse and zebrafish by the internet available tools. • Hints: protein sorting, translational modification, 3D structure……etc. 20