* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Protein Separation and Purification
Cell-penetrating peptide wikipedia , lookup
Molecular evolution wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Biochemistry wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein folding wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Chromatography wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein structure prediction wikipedia , lookup
Interactome wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein–protein interaction wikipedia , lookup
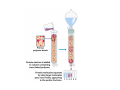
Protein Separation and Purification Methods rely on specific properties of protein Why purify a protein? Isolate Allows: Analysis of the biological properties Understand its structure Study interactions No single procedure can be used to isolate every protein Exploit specific characteristics (structure or function) of the protein. Different steps should exploit a different characteristic Ensure method has little/no effect on function Early steps involve releasing your protein from the cells (normally by homogenisation) and low resolving procedures to remove the bulk of the unwanted proteins Lysozyme Sonicator French Press Removing crude extract Ammonium sulphate precipitation (40%) exploits changes in the solubility of proteins as consequence of a change in ionic strength (salt conc.) of the solution At low salt, the solubility of a protein increases with salt concentration, ‘SALTING IN’. But as salt conc. (ionic strength) is increased further, the solubility of the protein begins to decrease, until a point where the protein is precipitated from solution, ‘SALTING OUT’. Low salt Strong attractive force + + + + + + - + - - - + - + Optimal salt conc. for solubility + + Weak attractive force + + + + + + - - + + + + + + High Salt Compete for water ++ + + -- -- + ++ + ++ + + + + -+ +- - -- - + + + - -+-+ + + + + - + -++ -++- + + - - -++ + + -++ + + + + + + + + + + + + -+-+-+ ++ ++ + + + + + + + +- + +++ - -+- + + + + + + + + + + - - +-+- - -++- - - -- - ++++ ++++ + -- - Debye-Huckel Theory Ammonium sulphate precipitation Hemoglobin Myoglobin Log of Solubility g mL-1 Fibrinogen 1 Serum albumin 2 Molarity of AmSO4 3 1M 2M AmSO4 3M The concept of a Column Ion Exchange Chromatography (IEC) Separates molecules based on their charge The side-chain groups of some amino acids are ionizable, e.g., lysine, arginine, histidine, glutamic acid, aspartic acid as are the N-terminal amino and C-terminal carboxyl groups Thus proteins are charge molecules and can have a different charge at a given pH because they have different compositions of ionizable amino acids For any given amphoteric protein, there will be a pH at which its overall charge is 0 (No. of negative charges equals the No. of positive charges) This is referred to as the ISOELECTRIC POINT (pI) or ISOTONIC POINT of the protein At a pH above its pI a protein will have a net negative charge while At a pH below its pI a protein will have a net positive charge IEC resins are made by covalently attaching Negatively or Positively charged functional groups to a solid support matrix to yield Cation or Anion exchangers, respectively Negatively charged exchangers bind positively charged ions – cations Positively charged exchangers bind negatively charged ions – anions At a pH=pI of a protein, it will not bind to an ion exchange resin When a charged molecule is applied to an exchanger of opposite charge, it is absorbed, while neutral ions or ions of the same charge are eluted in the void volume of the column (the volume that is not bound). The bound protein displaces the counterions Adsorbed molecules are commonly eluted with salt (changes ionic strength of the column buffer) or pH, which change the affinities of the bound proteins for the exchanger Gel Filtration Chromatography (GFC) GFC (also Size Exclusion Chromatography, Molecular Sieve Chromatography or Molecular Exclusion Chromatography) Separates molecules based on their size (& shape) It can also be used to determine the size and molecular weight of a protein Separation occurs due to the differential diffusion of various molecules into gel pores in a porous matrix. For protein purification, the matrix typically consists of porous beads (with pores of a specific size distribution) of an inert, highly hydrated gel Largest MW comes off first Separation is due to exclusion or inclusion from the gel matrix Small molecules diffuse into the gel pores, retarding their flow through the column, while large molecules do not enter the pores and are rapidly eluted from the column Proteins elute from the column in order of decreasing molecular weight Common gel matrices are dextran, agarose and polyacrylamide. These matrices are manufactured with different degrees of porosity, and thus can fractionate different size ranges of proteins Other Purification Methods: Affinity Chromatography: Separates molecules based on specific interactions between the protein of interest and the column matrix E.g. Antibodies which bind Protein Enzyme which binds a co-enzyme or inhibitor A ligand is covalently bound to a solid matrix (usually agarose) which is then packed into a chromatography column When a mixture containing the protein of interest is applied to the column, the desired protein is bound by the immobilised ligands, while all other proteins in the mixture, which should have no affinity for the ligand pass through and are discarded Affinity chromatography (with HIS-tagged proteins) Affinity chromatography can be performed using a number of different protein tags. poly-hisitidine The histidine tag is very short (6 His residues) Should not alter the conformation of the tagged protein Should not be involved in artificial interactions. The poly-his tag binds to a nickel chelate resin Eluted by 1.0 M imidazole List of Other Methods: Hydrophobic Interaction Chromatography: -hydrophobic interactions under high salt Chromatofocussing: -Fractionates based on a pH gradient generated in an ionexchange column HPLC: -Similar to IEF and GFC, but uses pressure to increase flow rates in columns with small particle size to increase resolution of peaks Methods for Assessing Protein Purity SDS-PAGE – Commonest method, rapid and sensitive SDS sodium dodecyl sulphate PAGE polyacrylamide gel electrophoresis Migration of a molecule in a electric field Separates materials based on size Isoelectric focusing (IEF) can also be used, which separates proteins by charge differences (induced by a pH gradient) 2D gel electrophoresis – Combination of SDS-PAGE and IEF Separates by charge in the first dimension and then by size in the second dimension Mass spectrometry Protein Purification Objectives Purity Stable Cost Time Protein purification steps Extraction Cell breakage chemical physical Debris removal straining centrifugation filtration Solubilization Inclusion bodies Urea Inhibition of proteases Preliminary concentration ammonium sulfate precipitation ultrafiltration dialysis Purification steps Ion exchange Affinity chromatography Gel filtration Preparative HPLC Final step Crystallization