* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chapter 11
Cre-Lox recombination wikipedia , lookup
Polyadenylation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Non-coding RNA wikipedia , lookup
Transcriptional regulation wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Expression vector wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene regulatory network wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA interference wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene expression wikipedia , lookup
List of types of proteins wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
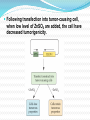
Nucleic Acids as Therapeutic Agents Nucleic Acids as Therapeutic Agents Many human disorders often results from the overproduction of a normal protein. Single stranded oligonucleotides could be used to hybridize to genes (antigene) or mRNAs (antisense) to reduce transcription or translation thereby reducing protein production. Synthetic RNA/DNA molecules called aptamers bind to proteins and prevent them from functioning. Ribozymes can be engineered to target mRNA. RNAi may be used instead of antisense RNAs or ribozymes. Inhibition of translation of specific mRNAs by antisense nucleic acid molecules. Antisense RNA: This is the backwards compliment of the normal RNA for a target gene. It is designed to bind the target mRNA and prevent translation. Demonstration is done in maligant glioma and prostate carcinoma cells which result from over production of insulin-like growth factor 1 and insulin-like growth factor 1 receptor, respectively. In both cases, cultured cells make large tumors when injected into rats. Cultured cells transfected with the proper antisense gene in an expression vector do not develop tumors. In both cases, the expression of the antisense cDNA is controlled by a Zn sensitive promoter from a metallothionein gene. Expression is turned on by presence of ZnSO4. Following transfection into tumor-causing cell, when low level of ZnSO4 are added, the cell have decreased tumorigenicity. Antisense oligonucleotides: short sequences designed to hybridize with mRNAs and prevent translation. Must hybridize to the target mRNA, be resistant to degradation, and be delivered into cells easily. Usually 15-24 nucleotides long. Can be designed from nearly any portion of the mRNA (5’ to 3’ ends of mRNAs, intron-exon boundaries, and regions that form stem loops have all been effective.) Proteomic analysis of cellular proteins can be used to determine if the protein production is reduced. The oligodeoxynucleotides are susceptible to degradation by intracellular nuclease. Various modifications have been tried to make the oligonucleotides more resistant to degradation without affecting the ability of antisense to hybridize to the target sequence. Modification includes adding sulfur to the phosphodiester backbone (phosphorthioate). The RNA-DNA duplex activates the Rnase H which cleaves hybrid molecules. It is considered to be “first generation” therapeutic agents. The second generation antisense therapeutic agents typically contain alkyl modifications at the 2’ position of the ribose. It is less toxic and more specific than phosphorothioate-modified molecules. The third generation antisense oligonucleotides contain a variety of modifications within the ribose ring, and/or the phosphate backbone, as well as being less toxic. Some modifications are made to both enhance stability and facilitate the binding to the target site. Free antisense oligonucleotides are not easily internalized by cells. Delivery is often accomplished using liposomes to facilitate cellular uptake. Utility of antisense oligonucleotides has been demonstrated in various cases. Narrowing stenosis of coronary arteries: alleviated by angioplasty but recurs quickly in 40% of patients because of a healing reaction. When antisense oligonucletides that targeted mRNA for proteins essential for cell cycle were applied to rats, restenosis was reduced by 90%. Smooth muscle cell proliferation implicated in atherosclerosis, hypertension, diabetes mellitus, and the failure of coronary bypassed grafted are presumably controlled by similar antisense therapeutics. Correction of a mutant splice site with an antisense oligonucleotide can be used to treat β-thalassemia. Ribozymes: naturally occurring catalytic RNA molecules that are ~40-50 nucleotides in length with separate catalytic and substrate binding domains. Comparing to protein therapeutics, it is less likely to evoke an immune response. The substrate-binding site combines by complementarity. The catalytic portion cleaves the target RNA at a specific site, thereby protein production is reduced. By altering the sequence, a ribozyme can be engineered to cleave any mRNA sequence. Representation of hammerhead (A) and hairpin (B) ribozyme-mRNA substrate complexes. Ribozymes may also be delivered directly by injection or liposomes, but must be chemically modified to avoid quick degradation. Also being tested for utility in fighting viral infections and prevent the accumulation of chemical/antibiotic resistance. Attempting to make synthetic DNA enzymes (deoxyribozymes) because DNA is much more stable. Aptamers are nucleic acid sequences, RNA or DNA, that bind tightly to proteins, amino acids, drugs, or other molecules. They are usually 15-40 nucleotides long, have highly organized secondary and tertiary structures, and bind with high affinity. The advantages are their high specificity, relative ease of production, low or no immunogenicity, and long-term stability. Aptamers are typically selected by a procedure known as SELEX (systematic evolution of ligands by exponential enrichment.) Interfering RNAs (RNAi): Adding dsRNA versions of a gene to a cell reduces the expression of the native gene by gene silencing. Occurs as a natural mechanism in many organisms where it is thought to be involved in protection from viruses and transposons. Upon introduction od dsRNA into a cell, Dicer complex binds to the RNA and cleaves it into an siRNA containing ~21 bp. The siRNA becomes part of RISC, directing the cleavage of the complementary mRNA. RNAi RNAi Antibody genes approach came from the observation that rainbow trout could be protected against hemorrhagic septicemia virus by passive immunization. Instead of administering a purified antibody, it is possible to inject an animal with DNA encoding the antibody. The cloned gene-vector constructed was injected into the circulatory system of rainbow trout, survival increased when challenged. The next step would be to create transgenic fish with antibody genes that confer passive immunity to various diseases. The effectiveness of any therapeutic agent depends upon the ability to deliver that agent to tissues where it is required. Systemic introduction often leads to the accumulation in tissues where the agent is not required and sometimes results in serious side effects. Viral vectors that deliver small nucleic acids to specific cellular targets have been developed. However, there are some safety concerns about using viral vector. There are several methods that have been developed. Intravenous injection Local injection at the site of pathology Packaging into cationic liposomes Physical methods Conjugated to another molecule There are a number of concerns about human gene therapy. How will the cell of target for correction be accessed? How will the gene be delivered? What proportion of the target cells must acquire the input gene to counteract the disease? Does transcription of the input gene need to be precisely regulated to be effective? Will it cause alternative physiological problems? Will it be maintained or repeated treatments be required. The viral vector are replication defective because they do not have a pol gene. Despite the advantages of viral vectors, they are often immunogenic, costly to maintain, and difficult to produce on a large scale without high-level expertise. The least complicated non-viral gene delivery system is the introduction of naked DNA. To avoid degradation of introduced DNA, DNAmolecular conjugates ha.e been developed. However, there are two major limitations. The frequency of transfection is often too low to be effective. The duration of gene expression is too brief. With the cell receptor-binding sites arrayed on the outside of the DNA-molecular conjugate, the complexes bound specifically to the specified cells . A human artificial chromosome (HAC) as therapeutic vector. Pros The DNA carrying capacity would be very large. This type of vector should have long-term stability and sustained expression. However, there are some issues must be addressed. Will HACs be efficiently introduced into the nuclei of target cells? Will effective levels of therapeutic gene expression be maintained for extended periods of time? The efficacy of using a particular gene along with a specific delivery system is tested on a small animals, typically mice to ensure not only the efficacy but also no unexpected side effects. A conjugate of cholesterol and an siRNA to mediate the uptake of siRNA into cells. Study are underway to improve the delivery of siRNA to treat various diseases. Use of a nonpathogenic strain of E. coli to deliver siRNAs to certain tissues. Negatively charged siRNA bind to positively charged atelocollagen. The complex greatly facilitates the delivery of siRNAs to specific tissues. A single-chain Fab fragment is fused to the positively charged polypeptide protamine, which binds to negatively charged siRNA. The Fab fragment acts to deliver the siRNA to specific cells. Since aptamers also bind specifically to a target protein, they can be used for targeting system.