* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Sequence-based prediction of protein interaction
P-type ATPase wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Phosphorylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein design wikipedia , lookup
Protein folding wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein domain wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Proteolysis wikipedia , lookup
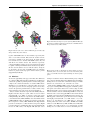
BIOINFORMATICS ORIGINAL PAPER Vol. 25 no. 5 2009, pages 585–591 doi:10.1093/bioinformatics/btp039 Sequence analysis Sequence-based prediction of protein interaction sites with an integrative method Xue-wen Chen1,2,∗ and Jong Cheol Jeong1 1 Bioinformatics and Computational Life Sciences Laboratory, Information and Telecommunication Technology Center and 2 Department of Electrical Engineering and Computer Science, University of Kansas, Lawrence, KS 66045, USA Received on June 2, 2008; revised on January 14, 2009; accepted on January 15, 2009 Advance Access publication January 19, 2009 Associate Editor: Limsoon Wong ABSTRACT Motivation: Identification of protein interaction sites has significant impact on understanding protein function, elucidating signal transduction networks and drug design studies. With the exponentially growing protein sequence data, predictive methods using sequence information only for protein interaction site prediction have drawn increasing interest. In this article, we propose a predictive model for identifying protein interaction sites. Without using any structure data, the proposed method extracts a wide range of features from protein sequences. A random forest-based integrative model is developed to effectively utilize these features and to deal with the imbalanced data classification problem commonly encountered in binding site predictions. Results: We evaluate the predictive method using 2829 interface residues and 24 616 non-interface residues extracted from 99 polypeptide chains in the Protein Data Bank. The experimental results show that the proposed method performs significantly better than two other sequence-based predictive methods and can reliably predict residues involved in protein interaction sites. Furthermore, we apply the method to predict interaction sites and to construct three protein complexes: the DnaK molecular chaperone system, 1YUW and 1DKG, which provide new insight into the sequence– function relationship. We show that the predicted interaction sites can be valuable as a first approach for guiding experimental methods investigating protein–protein interactions and localizing the specific interface residues. Availability: Datasets and software are available at http://ittc.ku.edu/~xwchen/bindingsite/prediction. Contact: [email protected] Supplementary information: Supplementary data are available at Bioinformatics online. 1 INTRODUCTION Protein–protein interaction plays an essential role in nearly all cell functions, such as promoting chemical reactions and acting as antibodies. Consequently, identification of protein interaction sites is critical for understanding protein function and for elucidating metabolic and signal transduction networks. It could also help in rational drug design studies (Gallet et al., 2000). A commonly used technique in identifying protein interaction sites is in silico ∗ To whom correspondence should be addressed. methods, which is of great importance in molecular recognition and are considered as a good starting point to form hypotheses in searching for potential pharmacological targets for the design of drugs (Gallet et al., 2000). Roughly speaking, computational methods can be categorized into two groups: molecular docking of two proteins with known structures and the identification of putative interaction sites on an isolated protein without knowing the structure of its partner or complex (Gallet et al., 2000). While a number of computational methods for predicting protein interaction sites have been developed over the years, most of them require known protein structure information (Aytuna et al., 2005; Bradford and Westhead, 2005; Chen and Zhou, 2005; Chung et al., 2006; Fariselli et al., 2002; Gabb et al., 1997; Helmer-Citterich and Tramontano, 1994; Jiang and Kim, 1991; Jones and Thornton, 1997a, b; Katchalski-Katzir et al., 1992; Keskin et al., 2005; Kuntz et al., 1982; Norel et al., 1995; Palma et al., 2000; Salemme, 1976; Shoichet and Kuntz, 1991; Walls and Sternberg, 1992; Warwicker, 1989; Wodak and Janin, 1978; Zhou and Shan, 2001). Despite much effort in structural genomics, the amount of protein structures, determined by timeconsuming and expensive experimental technologies, is significantly smaller than those of protein sequences produced by large-scale DNA sequencing methods. For example, by July 29, 2008, there are 392 667 identified protein sequences in Uniprot/Swissprot (reviewed, manually annotated) (Uniprot, 2008) and only 47 978 known protein structures in PDB (Berman et al., 2000). Thus, it is now more important than ever to identify protein interaction sites from amino acid sequences only, without knowing structural data. There are several studies attempted to address the sequence-based interaction site prediction problem. Kini and Evans (1996) observed that proline is the most common residue in a large number of protein interaction sites. Pazos et al. (1997) used multiple sequence alignment to detect correlated changes to a group of interacting protein domains for predicting contacting pairs of residues. Gallet et al. (2000) analyzed hydrophobicity distribution and amino acid frequencies in known interaction sites for identifying linear stretches of sequences. Most recently, more complicated machine learning methods are applied to predict interaction sites. Yan et al. (2003) applied support vector machines (SVMs) to predict interface sites with features extracted from sequence neighbors for each target residue. Wang et al. (2006) also employed SVMs as classifiers with features extracted from spatial sequence and evolutionary conservation scores based on a phylogenetic tree. © The Author 2009. Published by Oxford University Press. All rights reserved. For Permissions, please email: [email protected] 585 X.-w.Chen and J.C.Jeong Sequence-based method, increasingly important in protein interaction site prediction, is still in its infancy. Several issues exist that make the prediction from sequences a very difficult task. The two main problems are: (i) the biological properties that are responsible for protein–protein interactions are not fully understood, which leads to the difficulty of extracting informative features common to all the binding sites; and (ii) the number of interacting sites of a protein is much smaller than that of non-interacting sites, which leads to a very challenging problem, the so-called imbalanced data classification problem. Our article addresses these problems by extracting a wide variety of features from amino acid sequences and by developing a random forestbased method to effectively integrate these features and at the same time deal with imbalanced-data problems. The extracted features can be grouped into three categories: physicochemical properties and evolutionary conservation score, residue-based distance matrix and sequence profile. While each group of features may represent common characteristics for a certain number of interaction sites, none of the features is the dominant factor that is capable of describing the common effect among all the interface residues. For example, hydrophobicities may be useful for predicting interaction sites in homodimers, they are, however, of moderate power to predict interaction sites in other type of complexes (Jones and Thornton, 1996; Lo Conte et al., 1999). Thus, effectively integrating a large number of features is critical for a reliably predictive model. To utilize all the extracted features, an integrative random forest framework is developed. Our results evaluated on 99 polypeptide chains show that the proposed method outperforms two other sequence-based methods. Furthermore, we apply this method to identify potential interaction sites for three protein complexes: the DnaK molecular chaperone system, 1YUW and 1DKG, which may provide new insight into the sequence–function relationship. 2 2.1 METHODS Extracting a large number of features To build a predictor that can distinguish interface residues from noninterface sites, we extract features based on physicochemical property, evolutionary conservation score, amino acid distances, and position-specific score matrix (PSSM). Instead of using one long feature vector, we divide the features into three groups based on their sources, as features extracted from different sources may have different distribution (e.g. hydrophobicity in the physicochemical feature set versus the shortest distance of amino acid residues to a target residue in distance features) (see Supplementary Material for detailed discussions). Group 1—physicochemical features and evolutionary conservation score: the first two features are hydrophobicity and hydrophobic moments, which were initially used to distinguish membrane α-helix proteins from soluble proteins (Eisenberg et al., 1982, 1984) and later to predict protein binding sites in the apolipoprotein E sequence (De Loof et al., 1986; Gallet et al., 2000). For each amino acid, a sliding window centered at this amino acid is moved along the protein sequence and the mean hydrophobicity and mean hydrophobic moment are calculated as follows and assigned to the center amino acid. N 1 hn(i ) (1) < Hi >= 2N +1 n=−N < µHi >= 586 (i) (2N +1) is the size of the sliding window centered around amino acid i, hn is the hydrophobicity of the amino acid (AA) that is nAA’s away from the AA i, and δn is the gyration angle between two consecutive residues in the sequence. Gallet et al. (2000) found the method to be most successful when they used N = 5 and δn = 100˚. The hydrophobicities of each amino acid are taken from the scale developed by Eisenberg et al. (1984). We also extract seven other physicochemical properties including hydrophilicity, hydrophilic moment, propensity, propensity moment, isoelectric point, isoelectric moment and mass (Jones and Thornton, 1997a; Voet and Voet, 2004). The residue interface propensities quantify whether an amino acid is possibly exposed to solvent or buried in an interface (Jones and Thornton, 1997b). Furthermore, evolutionary conservation score for each residue is also calculated by HSSP (Schneider and Sander, 1996) and used as a feature. Thus, for each residue, we can extract nine physicochemical features and one evolutionary conservation score. Group 2—amino acid distance: the frequency of occurrence of proline residues was initially used for analyzing interaction sites by Kini and Evans (1996). They examined 1600 protein–protein interaction sequences and found proline residues on at least one side of 88.2% of the binding sites. The proline residues generally occurred within four residues of the binding site and often within two residues. Inspired by this idea, we examine the shortest distance from the current residue to 20 amino acid residues. This will create a vector with a size of 20 for each residue. Group 3—PSSM: PSSM is calculated by HSSP using multiple sequence alignment. The likelihood of 20 amino acid substitutions at a given alignment position is used as our PSSM features. For each target residue we are considering, its features are extracted by using a sliding window with a size of 21 centered on this target residue, i.e. the feature vector for the central residue consists of features extracted from itself and 10 amino acids on each side of this residue. Consequently, the total numbers of features for each target residue are 210, 420 and 420 for groups 1, 2 and 3, respectively. While each group of features may not be the dominant factor to describe the common effect among all the interface residues, collectively, they are capable of effectively characterizing protein interface sites. Since the total number of features is 1050, a carefully designed classifier is needed to effectively utilize the large size of features. Next, we describe the random forest method. 1 2N +1 2 N 2 12 N (i ) (i ) hn sin δn + hn cos (δn ) (2) n=−N n=−N 2.2 Constructing an integrative random forest model To effectively utilize the large number of extracted features and to deal with the imbalanced data classification problems, we herein describe an integrative random forest method for predicting interaction sites. Random forest tree has been applied to protein–protein interaction prediction in our recent work (Chen and Liu, 2005), but not to binding site problems. When the input space is extraordinarily large as in our application, random subspace feature selection introduced by Ho (1998) can improve classifier diversity. A random forest consists of an ensemble of decision trees from randomly sampled subspaces of the input features, and final classification is obtained by combining results from the trees via voting (Breiman, 2001). It is crucial to produce a large number of sufficiently different trees when using the combined power of multiple trees for increase in accuracy. The use of randomization in feature selection is a way to explore various possibilities of subspaces. While most classification methods suffer from the curse of dimensionality, the random subspace feature selection method can take advantage of the high dimensionality. In contrast to the Occam’s Razor, the method improves accuracy as it grows in complexity (Ho, 1998). The random forest can also deal with imbalanced data problems. It constructs many decision trees and each is grown from a different subset of training data. To construct individual decision tree, training samples are randomly selected with replacement from the original training dataset. In our application, to build each tree, we randomly select the same number of samples for each class, which converts the imbalanced data problem to multiple balanced data classification problems. If the number of positive samples (minority class) in the original training set is N, then N samples are Sequence-based prediction of protein interaction sites randomly drawn with replacement for each class. At each splitting or decision node, the best splitting feature is chosen from a randomly selected subspace of m features where m is much smaller than M total number of features. Each tree in the forest is grown to the largest extent possible without pruning. To classify a new object, each tree in the forest gives a classification which is interpreted as the tree ‘voting’ for that class. The final classification of the object is determined by majority votes among the classes decided by the forest of trees. Furthermore, for each group of features, we generate a random forest classifier. This is because features from different groups have different distribution. Building a forest classifier for each group of feature can effectively integrate all the features for better performance. For each feature group, we generate 100 trees: each tree is built using 100 randomly selected features from each feature group and the same number of positives and negatives. The final decision is made by majority vote. 3 EXPERIMENTAL RESULTS 3.1 Data sources The proteins used in this article were extracted from a set of 70 protein–protein heterocomplexes used in the studies of Chakrabarti and Janin (2002) and Yan et al. (2004). Redundant proteins and molecules with fewer than 10 residues and proteins with sequence identity ≥30% were removed. Some proteins which are not available in HSSP and DSSP programs (Kabsch and Sander, 1983) were also omitted. Finally, we end up with 54 heterocomplexes for our studies. Table 1 lists the 99 polypeptide chains extracted from the 54 heterocomplexes downloaded in PDB, which can be grouped into six categories: antibody–antigen, protease–inhibitor, enzyme complexes, large protease complexes, G-proteins and miscellaneous. Among 27 445 residues in the 99 polypeptide chains, we extract 13 774 surface residues based on their relative solvent accessible surface areas (RASA) calculated by the DSSP program: a residue is considered as a surface residue if its RASA is >25%. Furthermore, a surface residue is defined as an interface residue if the difference of accessible surface areas (ASA) between its unbound molecule and bounded complex is >1 Å2 . The definitions for surface residues and interface residues are commonly used in other literatures (Gong et al., 2005; Jones and Thornton, 1996; Jones and Thornton, 1997a; Nguyen et al., 2006; Rost and Sander, 1994; Wang et al., 2006; Yan et al., 2003). Among the 13 774 surface residues, 2829 residues are defined as interface residues (positive class). Thus, the number of non-interface residues including both non-binding surface residues and non-surface residues (negative class) is 24 616. Apparently, this is an imbalance data classification problem where the ratio of negative to positive samples is about 9:1. 3.2 Evaluation criteria To measure the performance of each predictor, we use leave-oneout cross-validation (LOOCV) and the following criterion functions, where true positive (TP) is the number of true interface residues that are predicted correctly; true negative (TN) is the number of true noninterface residues that are predicted correctly; false positive (FP) is the number of true non-interface residues that are predicted to be interface residues; and false negative (FN) is the number of true interface residues that are predicted to be non-interface residues. TN+TP Overall accuracy: TN+FP+FN+TP TP Sensitivity positive accuracy : FN+TP TN Specificity negative accuracy : FP+TN Balanced accuracy: Positive Accuracy×Negative Accuracy TP×TN−FP×FN TP+FN TP+FP TN+FP TN+FN Correlation coefficient (CC): The overall accuracy is the ratio of the number of correctly predicted residues (both positive and negative) to the total number of residues. It measures the overall performance of a classifier. In our application, since the number of positive samples is much smaller than that of negative samples, the overall accuracy may not be a good measure for evaluating the performance of a predictor. For imbalanced data classification, balanced accuracy and receiver operating characteristic (ROC) curves are typically used, where balanced accuracy is related to the product of both positive accuracy and negative accuracy and ROC curves are generated in terms of sensitivity and specificity. Additionally, CC, ranging from −1 to +1, is also a good measure. Its value is -1 for a worst possible predictor, +1 for a best possible predictor and 0 for a random predictor. 3.3 Leave-one-out test results To evaluate the performance, we compare the proposed method to two sequence-based methods. The first method, introduced by Yan et al. (2003) uses PSSM with 11 neighbor residues. The second method, proposed by Wang et al. (2006) uses PSSM and evolutionary conservation score with 11 neighbor residues. Both methods use support vector machines (SVMs) for prediction. We implement the same methods and procedures as described in Table 1. Protein categories and polypeptide chains with PDB ID Antibody-antigen Protease-inhibitor Enzyme Large-protease G-proteins Miscellaneous 1AO7_A, 1AO7_B, 1AO7_D, 1AO7_E, 1DVF_AB, 1DVF_CD, 1IAI_LH, 1IAI_MI, 1JH1_A, 1KB5_AB 1KB5_LH, 1NCA_LH, 1NCA_N, 1NFD_ABCD, 1NFD_EFGH, 1NMB_LH, 1NMB_N, 1NSN_LH, 1NSN_S 1OSP_LH, 1OSP_O, 1QFU_A , 1QFU_B, 1QFU_H, 1QFU_L, 1YQV_LH, 2JEL_LH, 2JEL_P, 3HFM_LH 1ACB_E, 1ACB_I, 1AVW_A, 1AVW_B, 1CHO_I, 1FLE_E, 1FLE_I, 1HIA_ABXY, 1HIA_IJ 1MCT_A, 1STF_E, 1STF_I, 1TGS_I, 1TGS_Z, 2SIC_I, 2SNI_E, 2SNI_I, 3SGB_E, 4CPA_I 1BRS_ABC, 1BRS_DEF, 1DFJ_E, 1DFJ_I, 1DHK_A, 1DHK_B, 1FSS_A 1FSS_B, 1GLA_F, 1GLA_G, 1UDI_E, 1UDI_I, 1YDR_E, 1YDR_I 1BTH_PQ, 1DAN_LH, 1DAN_TU, 1TBQ_LHJK, 1TBQ_RS, 1TOC_ABCDEFGH, 1TOC_RSTU, 4HTC_I 1AGR_AD, 1AGR_EH, 1GG2_A, 1GG2_B, 1GG2_G, 1GOT_A, 1GOT_B 1GOT_G, 1GUA_A, 1GUA_B, 1TX4_A, 1TX4_B, 2TRC_P 1AK4_AB, 1ATN_A, 1ATN_D, 1DKG_AB, 1EFN_AC, 1FC2_C, 1FC2_D, 1HWG_A 1HWG_BC, 1IGC_A, 1IGC_LH, 1SEB_ABEF, 1YCS_A, 1YCS_B, 2BTF_A, 2BTF_P 587 X.-w.Chen and J.C.Jeong 1 True positive rate (sensitivity) 0.9 0.8 0.7 0.6 0.5 0.4 0.3 Our-All Yan-All Wang-All 0.2 0.1 0 0 0.1 0.2 0.3 0.4 0.5 0.6 1-specificity 0.7 0.8 0.9 1 Fig. 2. Comparison of prediction performance in terms of the best balanced accuracy for three sequence-based methods. Fig. 1. The ROC curves for three sequence-based predictors. their papers. All the methods are trained and tested on the same datasets. To evaluate the performance, a LOOCV is used: each time, one of the 99 polypeptide chairs (including all the interface and noninterface residues in this polypeptide chain) is used as test data and the remaining 98 chains are used as training data; this process is repeated 99 times and the final results are averaged over the test results. Figure 1 shows the ROC curves for three sequence-based predictors. An ROC curve is a plot of the sensitivity versus (1 − specificity) for a binary classifier as its decision boundary is moved. Sensitivity measures the capability of predicting positive samples (interface residues) correctly and specificity determines if any non-interface residues are incorrectly predicted as interface residues. The ROC curve of our method is constructed by changing the threshold we place in the majority vote of decision trees. Typically, majority votes win where the threshold is zero. A threshold at five implies that at least five more votes of binding sites than these of non-binding sites are necessary to classify a residue as binding site. Otherwise, the residue is predicted as non-binding site. Therefore, with different thresholds, our model will produce different values of specificity and sensitivity. The ROC curve for Yan’s method is constructed by varying the threshold (bias) for a SVM decision boundary. Wang’s method consists of five SVM models. Thus, the ROC curve for Wang’s method is constructed by varying both the decision boundary of each SVMs with the same bias and the threshold for the majority vote of SVMs. The proposed method significantly outperforms Yan’s and Wang’s methods in terms of ROC curves: for example, with a specificity rate of 70%, the sensitivities of Yan’s, Wang’s and our methods are 30%, 39% and 73%, respectively. Another function we use is CC, which measures how predicted results correlate with actual data. The CC values range from negative one (worst possible prediction) to positive one (perfect prediction). For a random predictor, the CC value is zero. With a specificity rate of 70%, the CC values are 0.00, 0.06 and 0.28 for Yan’s, Wang’s and our methods, respectively. With a sensitivity rate of 70%, the CC values are 0.02, 0.05 and 0.28 for Yan’s, Wang’s and our methods, respectively. Thus, the proposed method outperforms other two methods and is significantly better than random guessing. We also compare the best results on balanced accuracies (the square root of product of positive accuracy and negative accuracy), which are commonly used for imbalanced data classification problems. Our method improves 23% and 17% in balanced accuracy compared with 588 (a) (b) Fig. 3. Predicted results of chains 1IAI_LH and 1IAI_MI in 1IAI using (a) our method, and (b) Wang’s method. Yan’s and Wang’s methods, respectively (Fig. 2). The results clearly demonstrate that the proposed method is capable of predicting protein interaction sites with significantly better performance than these previous sequence-based methods. We further examine the predicted results using Jmol (Jmol) and VMD software (Humphrey et al., 1996). For the results presented in Figures 3 and 4, the model is trained using training data and decisions are made without changing the threshold (e.g. in our method, simple majority vote is used). In Figures 3 and 4, each sphere represents an atom. Green sphere denotes true positives (true interface residues that are correctly predicted), blue sphere represents false negatives (interface residues that are predicted as non-interface residues) and red sphere indicates false positives (noninterface residues that are predicted as interface residues). Figure 3 shows the predicted interaction sites for four chains, L, H, M and I in idiotype-anti-idiotype Fab complex (Ban et al., 1994), using our method (Fig. 3a) and Wang’s method (Fig. 3b) obtained by leaving out each chain and training the predictive models on the remaining chains. Result from Yan’s method is similar to Figure 3b. Figure 4 shows the predicted interaction sites for two chains of 2CIO. Note that the structure of 2CIO was not available when we originally trained the model. Thus, it was not included in the group of 99 chains. We use 2CIO as a third, independent data for validating the three models. We also observe that our method identifies significantly more interaction residues for some complexes than the other two methods (more results can be found in the Supplementary Material: Figs S3 and S4). In conclusion, we showed that the proposed method outperformed two other sequence-based methods. To understand whether the improvement is due to the choice of the predictive model or the use of new features, we conducted tests using our random forest tree with the same PSSM features as those used in Wang’s method. The areas under ROC curve (AUC) for random forest and Wang’s classifier Sequence-based prediction of protein interaction sites (a) Fig. 5. DnaK molecular chaperone system: (a) DnaJ (PDB ID 1XBL), (b) the structure of DnaK C-terminal and (c) the structure of DnaK N-terminal. Orange structure denotes ATPase domain. (b) (c) Fig. 4. Predicted results of two chains in 2CIO using (a) our method, (b) Wang’s method and (c) Yan’s method. together with PSSM features are 0.75 and 0.58, respectively. The AUC of our method is 0.80. Thus, both the classifier and the new features contribute to the improvement: the AUCs obtained from random forest tree method with PSSM features only and with all the new features increase 0.17 and 0.22, respectively, compared with Wang’s method. As expected, random forest method is capable of dealing with imbalanced data classification problems. We also used the student’s t-test to rank the features: among the top 50 features, the best four are PSSM features, the remaining features are physicochemical properties and the distance profiles (Supplementary Table S4). 3.4 Fig. 6. Predicted results of 1YUW using our method. Black color denotes C-terminal (amino acid 395–554), blue color denotes N-terminal (amino acid 1–383) of DnaK, and atoms in predicted binding sites shown as purple spheres. Blind test To show the applicability of the proposed method, three blind tests are conducted. Without knowing the true binding sites, the blind tests evaluate the capability of predicting interface residues with peptide complexes. To construct 3D structures, we used the VMD software (Humphrey et al., 1996). Figures 5–7 show the results, where again, each sphere represents an atom and purple spheres denote these atoms in the predicted potential interface residues, and the orange spheres in Figure 7 denote the atoms also shown in Figure 5. First, we test three structural components of the DnaK (eukaryotic Hsp70) molecular chaperone system, which is used in another study (Fariselli et al., 2002). A chaperone system aids protein folding/unfolding. The first two components are two DnaK domains: a C-terminal domain (1DKX, PDB ID) and a N-terminal domain (1DKG, PDB ID), which are binding and releasing together. The third component is DnaJ (1XBL, PDB ID). The Hsp70 DnaK proteins are aided by the so-called J-domain cochaperones (Hsp40 proteins in eukaryotes, and DnaJ in prokaryotes), which dramatically increase the ATP activity of the Hsp70s. ATP hydrolysis causes locking in of substrates into the substrate-binding cavity of Hsp70 and cochaperone DnaJ modulates these ATP hydrolysis and substrate binding and is associated with conformational changes in DnaK (Gassler et al., 1998; Greene et al., 1998; Suh et al., 1998). DnaJ structures (PDB ID 1XBL) consist of four α-helices and a loop region containing HPD motif (tripeptide of histidine, proline, and aspartic acid residues) between two α-helices (Fig. 5a). This HPD motif is highly conserved and presented in almost all known J domains and this is critical to stimulate Hsp70 ATPase activity and mutations on the conserved tripeptide HPD of the J-domain abolish the ability of proteins to function with Hsp70 proteins; therefore, the HPD tripeptide could mediate specific interactions between Hsp40 and Hsp70 proteins (Fariselli et al., 2002; Gassler et al., 1998; Greene et al., 1998; Hennessy et al., 2000; Suh et al., 1998). Our method predicted two amino acid residues, 30-MET and 36-ARG which are near the HPD motif (33-HIS, 34-PRO, and 35-ASP). This is similar to the results of Greene et al. (1998) that the potential binding sites are residues between 1 and 35 and binding sites are 589 X.-w.Chen and J.C.Jeong reported binding regions II, III, IV, V and VI. We also predicted some binding sites in the upper right corner on chain B in GrpE, as a result of the fact that GrpE exists as a dimmer. 4 (a) (b) Fig. 7. Predicted results of three chains in 1DKG using our method. (a) DnaK N-terminal chain D of 1DKG, orange spheres are these atoms in these predicted interface residues shown in Figure 5c, and (b) GrpE, chain A and B of 1DKG. All spheres show our predicted atoms: purple spheres denote the atoms in the predicted interface residues and orange spheres denote the atoms also shown in Figure 5. concentrated on the outer surface of helix II which is a right-side α-helix where our prediction, 30-MET, is located; therefore, our predictions on DnaJ are reasonable. In addition 71-HIS and 74-PHE are also shown as conserved residues based on the consensus of amino acid position (Hennessy et al., 2000). For Hsp70 DnaK-Nterminal (PDB ID 1DKG) in Figure 5c, our prediction detected three residues (13-ASN, 116-SER, 174-ALA) on ATPase domain (orange color in Fig. 5c). Other researches (Davis et al., 1999; Gassler et al., 1998) showed that most of the mutants, which affect interaction with C-terminal domain, are located in the bottom of ATPase domain. Our predictions are spatially very close to those mutants observed in Davis and Gassler’s studies. For Hsp70 DnaK-Cterminal (PDB ID 1DKX) in Figure 5b, our method predicted 6-THR, 405-GLY, 447-ARG and 483-ILE as interface residues. Mutants observed by Davis and Montgomery are located in the loops on sandwich sub-domain which are close to peptide-binding site; therefore, our predictions are in agreement with their experimental results (Davis et al., 1999; Montgomery et al., 1999). We believe that the predicted binding will provide new insights into the interaction. Figure 6 shows the direct binding between DnaK C-terminal and N-terminal on the protein 1YUW in Bus Taurus (Jiang et al., 2005). Notice that 1YUW is different from 1DKX in Escherichia coli (Zhu et al., 1996) (also shown in Fig. 5) in terms of both Cterminal sequences and their binding sites. Our results show that most predicted interface residues are condensed in alpha helix of C-terminal, which are in agreement with these in Jiang et al. (2005). We also observe some predicted binding sites in N-terminal, which are not in the binding regions between C-terminal and N-terminal. Some of these may be the interaction sites between N-terminal and other chains (e.g. GrpE). Finally, we examined another DnaK effector GrpE by using 1DKG (Harrison et al., 1997) co-crystallized with 1DKG N-terminal and GrpE. GrpE is a nucleotide-exchange factor that binds substoichiometrically to the ATPase unit which is DnaK N-terminal. Figure 7a shows the predicted binding sites in DnaK N-terminal, which are very close to the reported binding residues in the regions III, V and VI (Harrison et al., 1997). In Figure 7b, the predicted interaction residues on chain A of GrpE are also located in the 590 CONCLUSIONS As genome-sequencing projects provide biologists with ready access to the rapidly increasing pool of protein sequences, there is a growing demand for developing advanced computational methods for predicting potential protein binding sites by using sequence information only. In this article, we demonstrate a predictive system that can reliably identify protein interface sites for protein complexes. The proposed predictive system is based on the analysis of protein sequence information, without knowing protein structures. A wide variety of physicochemical properties and sequence profiling properties are effectively integrated using a random forest tree framework. The predicted interaction sites can be valuable as a first approach for guiding experimental methods investigating protein–protein interactions and localizing the specific interface residues. We illustrate the usefulness of the proposed method for predicting putative binding sites for the DnaK molecular chaperone system, 1YUW and 1DKG. In our future work, we will evaluate the relative importance of the features, which should help to understand the underlining binding process. ACKNOWLEDGEMENTS We wish to thank Ozlen Keskin for helping us in modeling 3D protein structures with VMD software. We also thank the reviewers for their valuable suggestions. Funding: National Science Foundation award (IIS-0644366). Conflicts of Interest: none declared. REFERENCES Aytuna,A.S. et al. (2005) Prediction of protein-protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics, 21, 2850–2855. Ban,N. et al. (1994) Crystal structure of an idiotype-anti-idiotype Fab complex. Proc. Natl Acad. Sci. USA, 91, 1604–1608. Berman,H.M. et al. (2000) The protein data bank. Nucleic Acids Res., 28, 235–242. Bradford,J.R. and Westhead,D.R. (2005) Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics, 21, 1487–1494. Breiman,L. (2001) Random forests. Mach. Learn., 45, 5–32. Chakrabarti,P. and Janin,J. (2002) Dissecting protein-protein recognition sites. Proteins, 47, 334–343. Chen,H. and Zhou,H.X. (2005) Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins, 61, 21–35. Chen,X.W. and Liu,M. (2005) Prediction of protein-protein interactions using random decision forest framework. Bioinformatics, 21, 4394–4400. Chung,J.L. et al. (2006) Exploiting sequence and structure homologs to identify proteinprotein binding sites. Proteins, 62, 630–640. Davis,J.E. et al. (1999) Intragenic suppressors of Hsp70 mutants: interplay between the ATPase- and peptide-binding domains. Proc. Natl Acad. Sci. USA, 96, 9269–9276. De Loof,H. et al. (1986) Use of hydrophobicity profiles to predict receptor binding domains on apolipoprotein E and the low density lipoprotein apolipoprotein B-E receptor. Proc. Natl Acad. Sci. USA, 83, 2295–2299. Eisenberg,D. et al. (1982) The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature, 299, 371–374. Eisenberg,D. et al. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol., 179, 125–142. Sequence-based prediction of protein interaction sites Fariselli,P. et al. (2002) Prediction of protein–protein interaction sites in heterocomplexes with neural networks. Eur. J. Biochem.FEBS, 269, 1356–1361. Gabb,H.A. et al. (1997) Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol., 272, 106–120. Gallet,X. et al. (2000) A fast method to predict protein interaction sites from sequences. J. Mol. Biol., 302, 917–926. Gassler,C.S. et al. (1998) Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl Acad. Sci. USA, 95, 15229–15234. Gong,S. et al. (2005) A protein domain interaction interface database: InterPare. BMC Bioinformatics, 6, 207. Greene,M.K. et al. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA, 95, 6108–6113. Harrison,C.J. et al. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science, 276, 431–435. Helmer-Citterich,M. and Tramontano,A. (1994) PUZZLE: a new method for automated protein docking based on surface shape complementarity. J. Mol. Biol., 235, 1021–1031. Hennessy,F. et al. (2000) Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones, 5, 347–358. Ho,T.K. (1998) The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell., 20, 832–844. Humphrey,W. et al. (1996) VMD: visual molecular dynamics. J. Mol. Graph, 14, 33–38, 27–38. Jiang,F. and Kim,S.H. (1991) “Soft docking”: matching of molecular surface cubes. J. Mol. Biol., 219, 79–102. Jiang,J. et al. (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol. cell, 20, 513–524. Jmol. Jmol: an open-source Java viewer for chemical structures in 3D. Available at http://www.jmol.org. Jones,S. and Thornton,J.M. (1996) Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA, 93, 13–20. Jones,S. and Thornton,J.M. (1997a) Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol., 272, 121–132. Jones,S. and Thornton,J.M. (1997b) Prediction of protein-protein interaction sites using patch analysis. J. Mol. Biol., 272, 133–143. Kabsch,W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. Katchalski-Katzir,E. et al. (1992) Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl Acad. Sci. USA, 89, 2195–2199. Keskin,O. et al. (2005) Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol., 345, 1281–1294. Kini,R.M. and Evans,H.J. (1996) Prediction of potential protein-protein interaction sites from amino acid sequence. Identification of a fibrin polymerization site. FEBS Lett., 385, 81–86. Kuntz,I.D. et al. (1982) A geometric approach to macromolecule-ligand interactions. J. Mol. Biol., 161, 269–288. Lo Conte,L. et al. (1999) The atomic structure of protein-protein recognition sites. J. Mol. Biol., 285, 2177–2198. Montgomery,D.L. et al. (1999) Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol., 286, 915–932. Nguyen,N. et al. (2006) Protein-protein interface residue prediction with SVM using evolutionary profiles and accessible surface areas. In Proceedings of IEEE Symposium on Computational Intellegence Bioinformatics Computation Biology. pp. 1–5. Norel,R. et al. (1995) Molecular surface complementarity at protein-protein interfaces: the critical role played by surface normals at well placed, sparse, points in docking. J. Mol. Biol., 252, 263–273. Palma,P.N. et al. (2000) BiGGER: a new (soft) docking algorithm for predicting protein interactions. Proteins, 39, 372–384. Pazos,F. et al. (1997) Correlated mutations contain information about protein-protein interaction. J. Mol. Biol., 271, 511–523. Rost,B. and Sander,C. (1994) Conservation and prediction of solvent accessibility in protein families. Proteins, 20, 216–226. Salemme,F.R. (1976) An hypothetical structure for an intermolecular electron transfer complex of cytochromes c and b5. J. Mol. Biol., 102, 563–568. Schneider,R. and Sander,C. (1996) The HSSP database of protein structure-sequence alignments. Nucleic Acids Res., 24, 201–205. Shoichet,B.K. and Kuntz,I.D. (1991) Protein docking and complementarity. J. Mol. Biol., 221, 327–346. Suh,W.C. et al. (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl Acad. Sci. USA, 95, 15223–15228. Uniprot (2008) The Universal Protein Resource (UniProt). Nucleic Acids Res., 36, D190–D195. Voet,D. and Voet,J.G. (2004) Biochemistry. J. Wiley & Sons, Hoboken, NJ. Walls,P.H. and Sternberg,M.J. (1992) New algorithm to model protein-protein recognition based on surface complementarity. Applications to antibody-antigen docking. J. Mol. Biol., 228, 277–297. Wang,B. et al. (2006) Predicting protein interaction sites from residue spatial sequence profile and evolution rate. FEBS Lett., 580, 380–384. Warwicker,J. (1989) Investigating protein-protein interaction surfaces using a reduced stereochemical and electrostatic model. J. Mol. Biol., 206, 381–395. Wodak,S.J. and Janin,J. (1978) Computer analysis of protein-protein interaction. J. Mol. Biol., 124, 323–342. Yan,C. et al. (2003) Identification of surface residues involved in protein-protein interaction-a support vector machine approach. In Proceedings of the Conference on Intellegence System Design Application. pp. 53–62. Yan,C. et al. (2004) A two-stage classifier for identification of protein-protein interface residues. Bioinformatics, 20(Suppl. 1), i371–i378. Zhou,H.X. and Shan,Y. (2001) Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins, 44, 336–343. Zhu,X. et al. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science, 272, 1606–1614. 591