* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Ch. 20 Tricarboxylic acid cyle Student Learning Outcomes
Proteolysis wikipedia , lookup
Biochemical cascade wikipedia , lookup
Photosynthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Biosynthesis wikipedia , lookup
Mitochondrion wikipedia , lookup
Microbial metabolism wikipedia , lookup
Electron transport chain wikipedia , lookup
Biochemistry wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
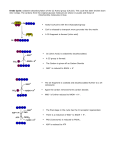
Chapt. 20 TCA cycle Overview TCA cycle Ch. 20 Tricarboxylic acid cyle Student Learning Outcomes: • Describe relevance of TCA cycle • Acetyl CoA funnels products • Describe reactions of TCA cycle in cell respiration: 2C added, oxidations, rearrangements-> NADH, FAD(2H), GTP, CO2 produced • Explain TCA cycle intermediates are used in biosynthetic reactions • Describe how TCA cycle is regulated by ATP demand: ADP levels, NADH/NAD+ ratio II. Reactions of TCA cycle • 2C unit Acetyl CoA • Adds to 4C oxaloacetate • Forms 6C citrate • Oxidations, rearrangements -> • Oxaloacetate again • 2 CO2 released • 3 NADH, 1 FAD(2H) • 1 GTP TCA cycle Reactions. A. Formation, oxidation of isocitrate: 2C onto oxaloacetate (synthase C-C • 2 C of Acetyl CoA are oxidized to CO2 (not the same 2 that enter) • Electrons conserved through NAD+, FAD -> go to electron transport chain • 2.5 ATP/NADH; 1.5 ATP/FAD(2H) • Net 10 high-energy P/Acetyl group Fig. 1 TCA cycle reactions Reactions of TCA cycle: • 1 GTP substrate level phosphorylation: TCA cycle (Kreb’s cycle) or citric acid cycle: • Generates 2/3 of ATP Fig. 3** synthetases need ~P) Aconitrase move OH Fig. 2 (will become C=O) Isocitrate Dehydrogenase oxidizes –OH, cleaves COOH -> CO2 also get NADH 1 TCA cycle reactions TCA cycle Reactions. B. α-ketoglutarate to Succinyl CoA: Oxidative decarboxylation releases CO2 Succinyl joins to CoA NADH formed TCA cycle reactions Fig. 3** TCA cycle Reactions. D. Oxidation of Succinate to oxaloacetate: Fig. 3** 2 e- from succinate to FAD-> FAD(2H) Fumarate formed H2O added -> malate 2 e- to NAD+ -> NADH Oxaloacetate restored GTP made from activated succinyl CoA (common series of oxidations to C=C, add H2O -> -OH, oxidize -OH to C=O) III. Coenzymes are critical: NAD+ III. Coenzymes are critical for TCA cycle • Many dehydrogenases use NAD+ coenzyme • NAD+ accepts 2 e- (hydride ion H-): -OH -> C=O • NAD+, and NADH are released from enzyme; • Can bind and inhibit different dehydrogenases • NAD+/NADH regulatory role (e-transport rate) Fig. 5 • FAD can accept e- singly (as C=C formation) • FAD remains tightly bound to enzymes Fig. 4 Fig. 6 membrane bound succinate dehydrogenase: FAD transfers e- to Fe-S group and to ETC 2 Coenzyme CoA in TCA cycle Coenzymes CoASH; TPP CoASH coenzyme forms thioester bond: • High energy bond Coenzymes CoASH, TPP (Figs. 8.11, 8.12) (Fig. 8.12 structure of CoASH formed from pantothenate) Fig. 7 Coenzymes in α-ketoacid dehydrogenase complex. C. α-ketoacid dehydrogenase complex: • 3 member family (pyruvate dehydrogenase, branched-chain aa dehydrogenase) α-ketoacid dehydrogenase enzyme complex: • 3 enzymes E1, E2, E3 • Coenzymes: TPP(thiamine pyrophosphate). Lipoate, FAD • Ketoacid is decarboxylated • CO2 released • Keto group activated, attached CoA • Huge enzyme complexes • (3 enzymes E1, E2, E3) • Different coenzymes in each Fig. 8 Fig. 9 3 Lipoate is a coenzyme Energetics of TCA cycle Energetics of TCA cycle: overall net -∆ ∆G0’ • Some reactions positive; • Some loss of energy as heat (-13 kcal) • Oxidation of NADH, FAD(2H) helps pull TCA cycle forward Lipoate coenzyme: • • • • • Made from carbohydrate, aa Not from vitamin precursor Attaches to –NH2 of lysine of enzyme Transfers acyl fragment to CoASH Transfers e- from SH to FAD Fig. 10 V. Regulation of TCA cycle Many points of regulation of TCA cycle: • PO4 state of ATP (ATP:ADP) • Reduction state of NAD+ (ratio NADH:NAD+) • NADH must enter ETC Fig. 12 Very efficient cycle: • Yield 207 Kcal from 1 Acetyl -> CO2 • (90% theoretical 228) • Table 20.1 Fig. 11 Table 20.2 general regulatory mechanisms Table 20.2 general regulation metabolic paths • • • • Regulation matches function (tissue-specific differences) Often at rate-limiting step, slowest step Often first committed step of pathway, or branchpoint Regulatory enzymes often catalyze physiological irreversible reactions (differ in catabolic, biosynthetic paths) • Often feedback regulation by end product • Compartmentalization also helps control access to enzymes • Hormonal regulation integrates responses among tissues: • Phosphorylation state of enyzmes • Amount of enzyme • Concentration of activator or inhibitor 4 Citrate synthase simple regulation Citrate synthase simple regulation: • Concentration of oxaloacetate, the substrate • Citrate is product inhibitor, competitive with S • Malate -> oxoaloacetate favors malate • • If NADH/NAD+ ratio decreases, more oxaloacetate If isocitrate dehydrogenase activated, less citrate Allosteric regulation of isocitrate Dehydrogenase Isocitrate dehydrogenase (ICDH): • Rate-limiting step • Allosteric activation by ADP • Small inc ADP -> large change rate • Allosteric inhibition by NADH • Reflect function of ETC Fig. 13 Other regulation of TCA Regulation of a-ketoglutarate dehydrogenase: • Product inhibited by NADH, succinyl CoA • May be inhibited by GTP • Like ICDH, responds to levels ADP, ETC activity VI. Precursors of Acetyl CoA VI. Many fuels feed directly into Acetyl CoA • Will be completely oxidized to CO2 Regulation of TCA cycle intermediates: • Ensures NADH made fast enough for ATP homeostasis • Keeps concentration of intermediates appropriate Fig. 14 5 Pyruvate Dehydrogenase complex (PDC) Regulation of PDC PDC regulated mostly by phosphorylation: • Both enzymes in complex • PDC kinase add PO4 to ser on E1 • PDC phosphatase removes PO4 Pyruvate Dehydrogenase complex (PDC): • Critical step linking glycolysis to TCA • Similar to αKGDH (Fig. 20.15) • Huge complex; • Many copies each subunit: • PDC kinase: (Beef heart 30 E1, 60 E2, 6 E3, X) • • inhibited by ADP, pyruvate Activated by Ac CoA, NADH Fig. 15 Fig. 16 TCA cycle intermediates and anaplerotic paths Anaplerotic reactions TCA cycle intermediates - biosynthesis precursors • Liver ‘open cycle’ high efflux of intermediates: • Specific transporters inner mitochondrial membrane for pyruvate, citrate, a-KG, malate, ADP, ATP. Anaplerotic reactions replenish 4-C needed to regenerate oxaloacetate and keep TCA cycling: • Pyruvate carboxylase • Contains biotin • Forms intermediate with CO2 • Requires ATP, Mg2+ (Fig. 8.12) • Found in many tissues Fig. 17 GABA Fig. 18 6 Amino acid degradation forms TCA cycle intermediates Key concepts Amino acid oxidation forms many TCA cycle intermediates: • Oxidation of even-chain fatty acids and ketone body not replenish Fig. 19 • TCA cycle accounts for about 2/3 of ATP generated from fuel oxidation • Enyzmes are all located in mitochondrial • Acetyl CoA is substrate for TCA cycle: • Generates CO2, NADH, FAD(2H), GTP • e- from NADH, FAD(2H) to electron-transport chain. • Enzymes need many cofactors • Intermediates of TCA cycle are used for biosynthesis, replaced by anaplerotic (refilling) reactions • TCA cycle enzymes are carefully regulated Nuclear-encoded proteins in mitochondria Nuclear-encoded proteins enter mitochondria via translocases: • Proteins made on free ribosomes, bound with chaperones • N-terminal aa presequences • TOM complex crosses outer • TIM complex crosses inner • Final processing Review question Succinyl dehydrogenase differs from other enzymes in the TCA cycle in that it is the only enzyme that displays which of the following characteristics? a. It is embedded in the inner mitochondrial membrane b. It is inhibited by NADH c. It contains bound FAD d. It contains fe-S centers e. It is regulated by a kinase • Membrane proteins similar Fig. 20 7