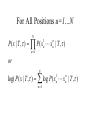* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Presentation Tuesday
Point mutation wikipedia , lookup
Public health genomics wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
Transposable element wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
Gene desert wikipedia , lookup
Maximum parsimony (phylogenetics) wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Human genome wikipedia , lookup
Metagenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Koinophilia wikipedia , lookup
Minimal genome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Pathogenomics wikipedia , lookup
Sequence alignment wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression programming wikipedia , lookup
Multiple sequence alignment wikipedia , lookup
Genome editing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
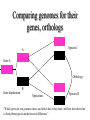
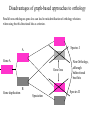
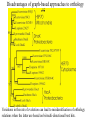
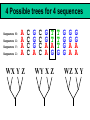
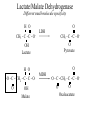
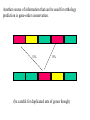
Orthology, and its relevance to protein function prediction Fitch 1970: “Where the homology is the result of gene duplication so that both copies have descended side by side during the history of an organism, (for example alpha and beta hemoglobin) the genes should be called paralogous (para= in parallel). Where the homology is the result of speciation so that the history of the gene reflects the history of the species (for example, alpha hemoglobin in man and mouse) the genes should be called orthologous (ortho=exact)” Comparing genomes for their genes, orthologs Species I A Gene A Orthologs Gene duplication B Speciation Species II “Which genes do two genomes share, and which don’t they share, and how does that relate to their phenotypical similarities and differences” Orthology: Who cares? Dystrophin related protein 2 Dystrophin Utrophin Dystrotelin Dystrobrevin The DYS-1 gene from C.elegans is not orthologous to dystrophin, that there is no effect of the knockout on the muscle cells is not so surprising. Best-bidirectional hits: a graph based approach Genome I 35% 23% 25% 30% Genome II Orthologs are expected to have relatively high levels of sequence identity to each other (compared to to other non-orthologous homologs), because they diverged relatively recently, and …… because they have similar functions…. (???) Large scale orthology determination is often done using bidirectional best hits An implementation of large-scale detection of pairwise orthology relations between genomes: 1) 2) Pairwise comparison of all genomes with each other, using the Smith-Waterman algorithm to detect “all” homologous relations (E < 0.01) between the predicted proteins. Select orthologous relations by selecting best-bidirectional hits between proteins. -Scoring as level of sequence identity * length of hit. -Including the possibility of gene fusion/fission (protein A from genome I can be orthologous to proteins B and C from genome II). By selecting genome I genome II A B C Genome I 35% 35% 25% 23% 25% Genome II 40% 30% 22% 20% 35% Genome III Multiple genomes can be used to check for consistency of bidirectional best hits. Solution to the non-transitivity of the concept of orthology sensu stricto is: “Group orthology” Conceptually: all proteins that are directly descended from one protein in the last common ancestor of the species one is interested in are considered orthologous to each other Operationally in a “graph-based approach”: Combine all connected “best triangular hits” into Clusters of Orthologous Groups (COGs, Tatusov et al, 1997). WWW.NCBI.NLM.GOV Gene duplications are creative, creating the possibility for developing new functions (in this case involved in carnitine synthesis) but …. They mess up orthology. Furthermore, intricate combinations of gene duplications and speciations make orthology nontransitive Inparalogs versus outparalogs: Inparalogs are due to relatively recent, species-specific gene duplications, e.g. Q9V6P0 and Q9VY24. Outparalogs are due to gene duplications that preceded speciations, e.g. Q9V6P0 vs. Q9VDM7 Disadvantages of graph-based approaches to orthology Parallel non-orthologous gene-loss can lead to misidentification of orthology relations when using best bi-directional hits as criterion. Species I A Gene A Gene loss Gene duplication B Speciation Non-Orthologs, although bidirectional best hits Species II Disadvantages of graph-based approaches to orthology Variations in the rate of evolution can lead to misidentification of orthology relations when the latter are based on bi/multi-directional best hits. Graph based approaches can recognize outparalogs as inparalogs (and vice versa) Because of independent loss events, and because of variable rates of evolution, in large gene families, orthology determination using bi/multi-directional best hits (graph-based approaches) does not always resolve separate orthologous and/or functional groups. One solution to this is the creation of phylogenies……… Prediction of orthology using phylogenies (unrooted) A tree-based approach would also allow a hierarchical, multilevel view on orthology, e.g. by including a numbering system Classic usage of phylogeny: inferring evolutionary history Dyall et al, Nature 2004 Hrdy et al, Nature 2004 How to make a tree 1) Distance based methods. Cluster the sequences based on relative levels of sequence similarity. Fast, but not a direct reconstruction of “what happened in evolution”. Neighbor Joining is the often used method here 2) Parsimony methods. Reconstruct the phylogeny that required the least amount of mutations. Slower (requires in principle the examination of all trees), but branch & bound makes it faster N=P I=3…..T(2i-5) and based on the questionable assumption that the least amount of events possible occurred in evolution. 3) Maximum likelihood methods. Find the phylogeny (including branch lengths) that, given a model of sequence evolution, was most likely to have produced to sequence alignment. This involves the comparison of all trees, estimating the branch lengths optimal for that tree, and subsequently estimating the likelihood of the complete tree . The slowest method of all, that furthermore requires knowledge of a large number of parameters Likelihood-Based Phylogeny 4 Possible trees for 4 sequences Sequence W: Sequence X: Sequence Y: Sequence Z: A A A A WX Y Z C C C C G G G A C C C C G G A A T T A G WY X Z T T T G G G G G G G A A G G A A WZ X Y All Possible Evolutionary Paths for one tree, for one collumn in the alignment T T A G A T G C A T G C A T G C Likelihood for One Path TTAG T G G L(path) = L(root) x P L(branches) =P(G) P(T|G)P(G|G) P(A|G)P(G|G) P(T|T)P(T|T) Calculating the likelihood of any path requires a model of sequence evolution (and an estimate of the time, and the mathematics to combine both) Kimura Jukes Cantor A a C A a a G a a a a 2a 2a T G General C A 2a 2a a b C a c T G d f e T The Jukes--Cantor Model, (including time…) αε αε αε 1 3α αε 1 3α αε αε S ( ) αε αε 1 3α αε αε αε αε 1 3α rt s S (t ) t st st st rt st st st st rt st st st st rt rt (1 3e 4t ) / 4 where st (1 e 4t ) / 4 Sum over all paths TTAG TTAG A A T T G G C A C T G C L(Column Cluster 1) = S L(all possible Evolutionary Paths) = L(path1) + L(path2) + L(path3) + … + L(path64) Likelihood of a phylogeny tree for one site x5 t4 x4 t2 t3 t1 x1 x2 x3 When x4 x5 are unknown, P( x1 x 2 x 3 | T , t ) 5 4 5 3 5 1 4 2 4 P ( x ) P ( x | x , t ) P ( x | x , t ) P ( x | x , t ) P ( x | x , t2 ) 4 3 1 x5 , x 4 Whole Sequence Likelihood WX Y Z L(Sequence) = L(each position i) i Choose the tree with the Maximum Likelihood. For All Positions u=1…N N P ( x | T , ) P ( x x | T , ) 1 u n u u 1 or N log( P( x | T , ) log P( x x | T , ) u 1 1 u n u Maximum Likelihood Phylogeny • Pick an Evolutionary Model (Modeltest can help) • For each position, Generate all possible tree structures • Based on the Evolutionary Model, calculate Likelihood of these Trees and Sum them to get the Column Likelihood for each OTU cluster. • Calculate Tree Likelihood by multiplying the likelihood for each position • Choose Tree with Greatest Likelihood Likelihood-based Phylogeny • Works well for distantly related sequences • Works well under different molecular clock theory • Can incorporate any desirable evolutionary model • Requires a good “model” of evolution • Requires fast computers Obtaining confidence in our tree Bootstrapping: 1) Generate e.g. 1000 alignments by random sampling (with replacement) from the real alignment. 2) Determine the phylogenetic tree for each alignment. 3) Count how often every subtree appears, and put those values at the internal branches of the complete tree. … bootstrap values tend to be on the conservative side…. Unrooted tree topologies A B A A B = C = C B D D C D = ((A,B),(C,D)) Bracket notation -Unrooted tree topologies only reflect relative evolutionary relations (In the primates the humans and chimpanzee are closer related to each other than either is to the the chimpanzee than they are to the Orang-Otang and the Gibbon) -Rooted trees reflect relative order of descendance (In the primates first the Gibbon branched off, then the Orang-Otang branched off, then the chimpanzee and then the humans) Orang-Otang Gibbon Chimp Human Orang-Otang Gibbon Baboon Chimp Human Rooting is important to separate different orthologous groups in a tree different functionalities. Berend’s lecture…. To construct a tree one needs a multiple sequence alignment: Different optimization criteria: Global optimization: The best would be to run multidimensional dynamic programming, which would be guaranteed to give you the optimal alignment (optimal here means: the best alignment given the similarity matrix and the gap penalties, not necessarily what is structurally the most likely course of events). Reconstructing evolution: Trying, along an evolutionary tree, to minimize the number of insertion/deletion events. Alignment Phylogeny Automatic sequence alignment use heuristics to obtain some kind of approximation of the optimal alignment. ClustalW: -pairwise sequence alignments of all pairs (N-1)^2 / 2. -select the most similar pairs of sequences, align those, and subsequently iteratively align the alignments. T_Coffee: -better but slower Muscle: Substrate specificities are not necessarily monophyletic (convergent evolution). Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Wu et al., PNAS 1999 Lactate/Malate Dehydrogenase Different small-molecule specificity H O CH3 - C - C - O- LDH CH3 - C - C - O- OH Lactate O Pyruvate H O O - C - CH2 - C - C OH Malate O- MDH -O - C - CH2 - C - C - O- O O -O O O Oxaloacetate Lactate/Malate Dehydrogenase H O CH3 - C - C - OOH Lactate negative -O H O - C - CH2 - C - C - O- O positive Arg 102 OH Malate Hannenhalli & Russell, JMB, 303, 61-76, 2000 Another source of information that can be used for orthology prediction is gene-order conservation. 35% 35% (be careful for duplicated sets of genes though) icd or leuB ? Prediction of orthology/function using a combination of sequence similarity and gene-order conservation Further reading • The quest for orthologs: finding the corresponding gene across genomes. Kuzniar A, van Ham RC, Pongor S, Leunissen JA. Trends Genet. 2008 Nov;24(11):539-51