* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Bottromycin wikipedia , lookup
Index of biochemistry articles wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Genetic code wikipedia , lookup
Interactome wikipedia , lookup
Biosynthesis wikipedia , lookup
Expanded genetic code wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein domain wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein adsorption wikipedia , lookup
Homology modeling wikipedia , lookup
Biochemistry wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
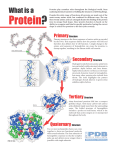
OSI Protein Modeling Challenge December 2010 Fall 2010 1 Joan Kiely, Stony Brook University [email protected] Debbie Pelio, Stony Brook University [email protected] http://www.stonybrook.edu/cesame Linda Padwa, Stony Brook University Kristen La Magna, Stony Brook University Shannon Colton, Ph.D., Technical Advisor Milwaukee School of Engineering http://cbm.msoe.edu/ Fall 2010 2 Protein Modeling Challenge • To compete successfully in the Protein Modeling Challenge, you will: – Meet and get to know Epidermal Growth Factor Receptor (a tyrosine kinase receptor), Tarceva and their roles in Lung Cancer – Build models that illustrate what you know about EGFR, Tarceva and Lung Cancer. – Become a maven of protein structure – Enhance your computer skills with Jmol Fall 2010 3 Web-Based Resources • This powerpoint presentation will serve as an interactive resource for your team to gain the knowledge they need to be successful in the Protein Modeling Challenge • You will find links distributed throughout this presentation, indicated by the blue underlined text • Follow these links to the appropriate sources • Good luck and have fun! Fall 2010 4 Protein Structure Resources • The following links will serve as tools to help you learn the basic information needed to be successful in this challenge. Please follow these links: – Basic Introduction to Protein Structure and Modeling – http://cbm.msoe.edu/stupro/so/index.html – Fall 2010 Protein Databank Molecule of the Month http://www.pdb.org/pdb/explore/motm.do 5 5 Protein Structure • • • Proteins are macromolecules Amino acids are the basic building blocks of proteins Working as a team, make an amino acid Sidechain (R-Group) Alpha-Carbon Carboxyl Group Nitrogen Amino Group • Fall 2010 s your amino acid L or D form? 6 6 D-Alanine Fall 2010 L-Alanine 7 Amino Acids Have Unique Chemical Characteristics • Each amino acid has the same “backbone” structure, but has different chemical groups (R groups or sidechains) attached • Working with another team, construct an amino acid and a dipeptide with a molymod kit NH2-CH-COOH R Fall 2010 8 8 Proteins Have Secondary Structure • A linear chain of amino acids is the protein “primary” structure • A chain of amino acids will spontaneously form stable “secondary structures”, ie: betasheet or alpha-helix • • Hydrogen bonds may stabilize these structures Science researchers would like to understand what controls this folding. Alpha Helix Beta-sheet – http://fold.it/portal/ Fall 2010 9 9 Proteins Fold Into a Tertiary Structure • Protein folding is due to the behavior of different chemical groups on amino acids in an aqueous environment • You can explore amino acid sidechain chemistry and protein structure through the game Fold It: http://fold.it/portal/ Fall 2010 10 10 Proteins Fold Into a Tertiary Structure • Proteins spontaneously fold into a specific three dimensional “tertiary” structure that governs a protein’s function Fall 2010 11 11 Protein Data Bank • The 3-dimensional structure of proteins is often determined by x-ray diffraction or NMR analysis • PDB file lists the X, Y, Z coordinates for each atom in a protein • Protein Data Bank http://www.pdb.org/pdb/home/home.do • PDB Molecule of the Month features the structure and function of a different protein each month http://www.pdb.org/pdb/static.do?p=education_discussion/ molecule_of_the_month/alphabetical_list.html Fall 2010 12 12 Molecule of the Month (MOM) • A monthly PDB feature written by David Goodsell http://www.pdb.org/pdb/static.do?p=education_discussion/molecule_of_the_m onth/alphabetical_list.html – Features a specific molecule – Describes protein function – Relates structure with function Epidermal Growth Factor June 2010 David Goodsell http://www.pdb.org/pdb/static.do?p=education_ discussion/molecule_of_the_month/pdb126 _1.html Fall 2010 13 13 Jmol • • • • Fall 2010 Jmol is a computer visualization software that displays data from a PDB file as a “3D” image of the molecule on the computer screen Jmol is Java-based and will work on most computers http://bioportal.weizmann.ac.il/oca-docs/fgij/index.htm http://cbm.msoe.edu/teachRes/index.html 14 14 Exploring Protein Structure with Jmol • Jmol allows you to identify elements of protein structure – Helix (magenta) – Sheet (yellow) – N-terminus (blue) – C-terminus (red) – Amino acid sidechains (CPK) – Alpha-carbon backbone model format Fall 2010 15 15 Mini-Toober Models (cont.) • Mark location of structures on Mini-Toober • Fold Mini-Toober into a 3D model representing protein Fall 2010 16 16 Protein Modeling Challenge • 2010 Event Rules www.stonybrook.edu/cesame • Pre-build model (40%) • On-site build (30%) • Written exam (30%) Fall 2010 17 17 EGFR Pre-Built Model 2010 • Epidermal Growth Factor and written description based on June 2010 Molecule Of the Month and Protein Databank File 1M17 residues 695-854 http://www.pdb.org/pdb/explore/motm.do – – Must arrive at Stony Brook by 4:30 December 1 for judging 40% team score Fall 2010 18 18 Written Exam 2010 • Exam covers material in: – PDB file 1M17 – Molecule of the Month article on Epidermal Growth Factor – Jmol – Campbell, Biology, will be used a the material base for questions on protein structure and function and cell communication – Taken as a group – Available at the exam will be: • • • PDB file, abstract, Molecule of the Month 30% team score Fall 2010 19 19 EGFR • • • On-site build PDB file will be provided on the day of the exam Students will build a portion of the EGFR receptor. They will be given: a toober, selected amino acid side chains, a computer, jmol and the pdb file. Fall 2010 20 20 Protein Modeling Challenge With National Science Content Standards • Science and Technology – – • Abilities of Technological Design Understandings about Science and Technology Life Science – The Cell – Physiology • Science as Inquiry – Abilities Necessary to do Scientific Inquiry • Physical Science – – Structure and Properties of Matter Chemical Reactions Fall 2010 21 21