* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download MCB Lecture 2 – Protein Metabolism
RNA polymerase II holoenzyme wikipedia , lookup
Interactome wikipedia , lookup
Citric acid cycle wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Paracrine signalling wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Messenger RNA wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Peptide synthesis wikipedia , lookup
Western blot wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Metalloprotein wikipedia , lookup
Gene expression wikipedia , lookup
Community fingerprinting wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Proteolysis wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transfer RNA wikipedia , lookup
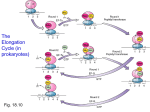
MCB Lecture 2 – Protein Metabolism What percentage of RNA is tRNA in the body? o 15% What percentage of RNA is rRNA in the body? o 80% What percentage of RNA is mRNA in the body? o 5% How many codons are there? o 64 What is the start codon? o AUG - Methionine What are the stop codons? o UAA – U Are Away o UAG – U Are Gone o UGA – U Go Away Wobble Positions: o What are the “normally” recognized codons? C=G A=U o What are the codons that untraditionally recognize two bases? U=A, U=G G=C, G=U o What recognizes three codons? What are the bases recognized? Inosine recognizes: I=A I=U I=C o What happens if there is an insertion or deletion one one nucleotide? There is a shift in the reading frame, so all of the amino acids are different. o What is a non-sense mutation? When a normal codon that codes for a particular amino acids is mutated and becomes one of the stop codons (UGA, UAA, UAG) Early termination of the strand o What is a silent mutation? When one of the nucleotides are changed but you still end up with the same amino acid (usually in the 3rd position) o What is a missense mutation? When one nucleotide is changed and it changes the amino acid placed in the chain (usually in the 1st position) o What type of mutation can lead to exon skipping or cryptic splicing? When the 5’ GU---AG 3’ has a mutation, the beginning and ending of exons can be misread and spliced out. Translation: How is the tRNA charged? o 1. ATP adds an AMP to the amino acid. o 2. AMP breaks off of amino acid. o Amino acid can now bind to the tRNA o Binds at the A residue of CCA (using free OH group of tRNA and carboxyl group of Amino Acid) What type of linkage is this? o Ester linkage How many ATP molecules are used when charging the tRNA? o 1, but 2 high energy bonds from it. Translation in Prokaryotes: Initiation: o What is the initiator tRNA first charged with? Formyl-Methionine o What is the enzyme that adds methionine to the tRNA? Synthetase o What is the enzyme that adds for formyl group to methionine? Transformylase The Prokaryotic Ribosome: o What Svedburg Number is the Large Ribosomal Subunit? 50S o What Svedburg Number is the Small Ribosomal Subunit? 30S o What Svedburg Number is the Combined Ribosomal Unit? 70S o What two rRNA’s make up the Large Ribosomal Subunit? 5S and 23S o What two rRNA’s make up the Small Ribosomal Subunit? 16S Initiation o What is the Shine-Dalgarno Sequence? A small sequence upstream of the AUG (start codon) where the 16S rRNA (small subunit) binds to initiate translation. o What is the A-Site? Acceptor site, where each new amino acids first attaches to the ribosome to be added to the strand. o What is the P-Site? Peptidyl transferase site, where the amino acid is added to the chain. Note that AUC (start codon) first binds here, NOT to the A-site like the rest of them. o What is the E-Site? Exit site, where the newly translated peptide chain leaves the ribosome. o Which rRNA subunit is a ribozyme (peptidyl transferase)? 23S rRNA in the 50S subunit (Large subunit) o What are the three prokaryotic translation initiation factors? IF1, IF2, IF3 o Which translation initiation factors bind first? Why? IF1: Binds to A-Site to prevent the use of the A-Site IF3: Binds to the edge of the 30S Small Subunit, preventing the 50S Large Subunit from binding prematurely. o What translation initiation factor binds next? Why? IF2: Brings in the fMET-tRNA to the P-Site. This requires GTP. o How is initiation completed? 1. The rMET-tRNA recognizes and binds to the AUG Codon at the P-Site. 2. 50S Subunit Hydrolyzes IF2 + GTP IF2 +GDP This causes all three IF’s to dissociate. Elongation: o What are the two factors involved in Elongation? EF-Tu EF-Ts EF-G o What is the first step of Elongation? Which factor is involved? tRNA that was already added is in the P-Site. EF-Tu + GTP brings a charges tRNA to the A-Site. o What are the bond forming steps of Elongation? What factor is involved? 1. The Tu protein gets hydrolyzed into Tu + GDP. 2. This allows it to let go of the charged tRNA, which then binds to the A-Site. 3. EF-Ts binds to EF-Tu to replace GDP with GTP so it can start over (recycles) o What type of bond is formed to elongate the strand? Peptide o Which two sites is the location of the formed peptide bond? A-Site and P-Site o Which ribosomal subunit catalyzes the reaction? 23S – Ribozyme with peptidyl transferase activity o Which factor is involved with translocation of the tRNA from the Psite to the E-site? TF-G o What releases the protein when it is done elongating? Release factors. o What is the Energetics of Translation Elongation? How many molecules are used? What are they used for? 1 ATP = 2 High Energy Bonds ATP: Charging tRNA AMP: Charging tRNA 2 GTP: 2 High Energy Bonds GTP: Binding charged tRNA to the A-Site using EF-Tu. GTP: Translocation Step: EF-G o For each Amino Acid incorporated, how many high-energy bonds are consumed? 4 Inhibitors of Prokaryotic Protein Synthesis: o How does Streptomycin inhibit Prokaryotic Synthesis? It binds to 30S (Small Subunit) and causes misreading of the genetic code and inhibits initiation of translation at higher concentrations. o How does Purinomycin inhibit Prokaryotic Synthesis? It is structurally similar to the 3’ end of aminoacyl tRNA, so it binds to the A-Site and participates in peptide bonding. This leads to a pre-mature ending of synthesis. o How does Tetracyclin inhibit Prokaryotic Synthesis? It blocks the A-site. o How does Chloramphenicol inhibit Prokaryotic Synthesis? It binds to the 50S (Large Subunit) and blocks bacterial Peptidyl Transferase. o How does Erythromycin inhibit Prokaryotic Synthesis? Binds to the 50S (Large Subunit) and blocks translocation. Eukaryotic Translation: What Svedburg Number is the Large Ribosomal Subunit? o 60S What Svedburg Number is the Small Ribosomal Subunit? o 40S What is the Svedburg Number for the entire Ribosome? o 80S What three rRNA’s make up the Large Subunit (60S) o 5S rRNA o 25S rRNA o 5.8S rRNA What one rRNA makes up the Small Subunit (40S) o 18S rRNA Orthologs for rRNA (Eukaryotes and Prokaryotes) What are the Eukaryotic Orthologs of these Prokaryotic rRNAs? o 5S rRNA 5.8S rRNA o 16S rRNA 18S rRNA o 23S rRNA 28S rRNA Which rRNA has no Ortholog? o The 5S rRNA in Eukaryotes has no ortholog in the Prokaryotes. Which part of the Ribosome has enzymatic activity? o The 28S rRNA in the 60S Eukaryotic Subunit (Large Subunit) Eukaryotic Translation: Initiation What is the Prokaryotic Ortholog of eEF1? What does it do? o EF-Tu. It brings an amino acid to the A-Site What is the Prokaryotic Ortholog for eEF2? What does it do? o EF-G. It does translocation. What does PAB bind to? o The 3’ Poly A Tail of mRNA What does eIF4E bind to? o The 5’ Cap of mRNA What does elF4G do? o Grabs the PAB and eIF4E, which brings together the two ends. What amino acid charges the first tRNA in Eukaryotes? o Methionine What factor binds to the Initiator Charged tRNA? o eIF2-GTP What general region does the Small Subunit binds before the Large Subunit is attached? o The Small Subunit Binds upstream from the AUG sequence and moves along until it finds the AUG. What happens once the AUG Start Site is found? o The GTP on eIF2-GTP is hydrolyzed. o Initiation Factors Dissociate o Large Ribosomal Subunit binds List the 6 steps of Eukaryotic Initiation of Translation: o 1. PAB binds to 3’ Poly A Tail o 2. eIF4E binds to 5’ Cap o 3. eIF4G grabs both eIF4E and PAB, and binds them together o 4. eIF2 + GTP (with met-tRNA) binds to the 5’ end of mRNA, upstream from AUG sequence. o 5. Locates AUG, and the subunit stops with tRNA in the P-Site o 6. 60S Subunit binds, allowing the eIF2 to dissociate and elongation can begin. Eukaryotic Translation: Elongation What factor brings in subsequent tRNA’s with Amino Acids bound? o eEF1. What site does the incoming tRNA bind to? o A-Site What factor induces translocation? How does it do it? What is it’s ortholog in Prokaryotes? o eEF2 induces translocation by binding to the A-Site and temporarily occupying it. The ortholog in Prokaryotes is EF-G. Which rRNA Subunit catalyzes the reaction? What is the bond called? What is the enzyme called? o 28S rRNA of the 60S Subunit, which forms a peptide bond via peptidyl transferase. How does translation terminate? o One of the stop codons is reached (UAA, UGA, UAG) and that signals release factors. Eukaryotic Toxins: What is Diptheria Toxin? What does it do? o It is an upper respiratory tract illness that causes significant neck swelling. o It inactivates eEF2 (which causes a stop of translocation) What is Ricin? What does it do? o Ricin is a toxic protein that inactivates the 28S Subunit, which stops peptidyl transferase activity. Protein Folding: What is co-translational protein folding? o Some proteins fold spontaneously into the correct conformation as they are being translated. What are heat shock proteins? o They are synthesized in large amount on heat exposure. o They have an affinity for exposed hydrophobic patches of incompletely folded proteins and use ATP to fold them correctly. What are the two types of heat shock proteins? o Hsp70 o Hsp60 What is Hsp70? o Acts co-translationally. o As protein is being translated, Hsp’s bind to the new strand and use ATP to fold it correctly. What is Hsp60? o Uses a “Chanel C’s” like complex that has hydrophobic binding sites to trap the incorrectly folded protein inside of the molecule. o A cap is added to the top (using ATP) and the protein inside gets folded correctly. o The cap leaves, and the correctly folded protein emerges. Post-Translational Modifications: What are terminal modifications? o The beginning and end of the protein is usually take off or modified What are signal sequences? o Signal sequences are areas at the beginning of a protein that can be cleaved off to create diversity. Which amino acids can be phosphorylated? o Ones with free –OH groups. o Examples: Serine, Threonine, Tyrosine What enzyme phosphorylates? o Kinase What enzyme dephosphorylates? o Phosphatase Does Kinase turn molecules off or on? o Both Kinase and Phosphatase can do either – off or on. Carboxylation is dependent on what? What bodily function does it relate to? o Carboxylation is dependent on Vitamin K. o Vitamin K leads to a modification of Glu and Gla, which is required for blood clotting. What Vitamin is essential for Hydroxylated Proline? What is Hydroylated Proline used for? o Vitamin C is essential for Hydroxylated Proline. o It is important for making collagen. Because collagen consist of repeating units Gly-X-Y, where X is usually Proline and Y is usually Hydroxylated Proline. o These units form strongly twisted alpha-chains that make collagen fibers. What residues have N-Linked Sugars? o Asn – (Asparagine) What residues have O-linked Sugars? o Ser and Thr (Serine and Threonine) What modifications can be made to Lysine? o Acetylated: The + is removed from the N o Methylated: Can have 1, 2, or 3 methyls attached, but N is still + There are three types of membrane protein attachments. What are they? o 1. Fatty acid is linked to N-terminal Glycine (N-C=O) o 2. Fatty acid is linked to Cysteine Residue (S-C=O) o 3. Fatty acid is linked to a Carboxy Terminal Cysteine (CH2-S-CH2) What are zymogens? What is an example? o Zymogens are inactive enzymes. (AKA Proenzymes) o Example: Inactive chymotrypsinogen is activated by Trypsin to make Active Trypsin Protein Degradation: What is Ubiquitin Ligase? What does it do? o It is a polyubiquitin chain on a lysine residue that directs a protein to the proteasome, so it will be degraded. Protein Folding Diseases: If a protein does not either: 1. Correctly fold without help. 2. Correctly fold with help of chaperone. Or 3. Incompletely fold and be degraded by the proteasome, what happens? o It becomes a protein aggregate (resistant to protease) What types of diseases does this cause? What are three examples? o Dominant diseases like Huntington’s, Parkinson’s, or Sickle Cell Anemia. What are Prion Diseases? o Prion diseases are caused by misfolding and aggregation of Prion Proteins. o Two Alpha Helices in the Prion Protein convert in four Beta Strands. o This results in Cross Beta Filaments, where the sheets are stacked. Why are Prion Diseases considered infectious? o Because once the Cross Beta Filaments form, they can stick to normal proteins and cause them to conform into the same, Beta Sheet conformation. What are three examples of Prion Diseases? o Creutzfeldt-Jacob Disease (CJD) in humans o Bovine spongiform encephalopathy (BSE) in cattle – mad cow o Scrapie in Sheep o ** These are caused by eating the meat of your own species**