* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chromosomal Inheritance - Bishop Seabury Academy
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genomic library wikipedia , lookup
Gene desert wikipedia , lookup
Oncogenomics wikipedia , lookup
Human genome wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Pathogenomics wikipedia , lookup
Public health genomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Ridge (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
DiGeorge syndrome wikipedia , lookup
Minimal genome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Y chromosome wikipedia , lookup
Neocentromere wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
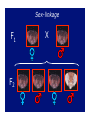
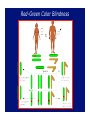
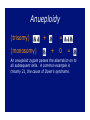
Chromosomal Inheritance 1. Explain how the observations of cytologists and geneticists provided the basis for the chromosome theory of inheritance. TT X tt 2. Describe the contributions that Thomas Hunt Morgan, Walter Sutton, and A.H. Sturtevant made to current understanding of chromosomal inheritance. Morgan P F1 X Sex-linkage F1 F2 X Sturtevant Linkage map of the pea genome (functional) 1% recombination frequency = 1 centimorgan Sutton In 1902, he provided sufficient evidence for the theory that genes are parts of chromosomes from direct observations on the behavior of chromosomes in reduction division. In 1903, he showed that the chromosomes behave by random segregation in the sex cells and recombination in fertilization, exactly as genes do. 3. Explain why Drosophila melanogaster is a good experimental organism. And, they reproduce prolifically every two weeks! 4. Define linkage and explain why linkage interferes with independent assortment. Gene linkage b = black body b+ = gray body vg+ = normal wings vg = vestigial wings b+b vg+vg bb vgvg X b+b vg+vg bb vgvg b+b vgvg bb vg+vg 575 575 575 575 b+b vg+vg bb vgvg X b+b vg+vg bb vgvg b+b vgvg bb vg+vg 965 944 206 185 5. Distinguish between parental and recombinant phenotypes. YyRr x yyrr yr YR Yr yR yr YyRr Yyrr yyRr yyrr Parental Types Recombinants 50% frequency of recombination b+b vg+vg bb vgvg X b+b vg+vg bb vgvg b+b vgvg bb vg+vg 575 575 575 575 b+b vg+vg bb vgvg X b+b vg+vg bb vgvg 1150 1150 Phenotypes Genotypes Expected results if genes are unlinked Expected results if genes are totally linked Actual Results Black body, normal wings bb vg+vg 575 Gray body, normal wings b+b vg+vg 575 1150 965 Black body, vestigial wings bb vgvg 575 1150 944 Gray body, vestigial wings b+b vgvg 575 206 185 Recombination frequency = 391 recombinants X 100 = 17% 2300 total offspring 6. Explain how crossing over can unlink genes. 7. Map a linear sequence of genes on a chromosome using given recombination frequencies from experimental crosses. Loci Recombination Frequency Approximate Map Units b vg 17.0% 18.5 cn b 9.0% 9.0 cn vg 9.5% 9.5 b cn 17 9 b cn vg 9.5 vg b 9 17 cn 9.5 vg The actual distance of 18.5 map units between b and vg is accounted for by the great distance between the loci and the potential for double crossovers that cancel each other out. 8. Explain what additional information cytological maps provide over crossover maps. Linkage map of the pea genome (functional) Cytogenetic map of the human genome (structural) 9. Distinguish between a heterogametic sex and a homogametic sex. The heterogametic sex (gender) produces two kinds of gametes and determines the sex of the offspring, the homogametic sex (gender) produces one kind of gamete. In humans, males are the heterogametic sex, while females are the homogametic sex. 10. Describe sex determination in humans. A British research team has identified a gene, SRY (sexdetermining region of Y), on the Y chromosome that is responsible for triggering the complex series of events that lead to normal testicular development, in the absence of SRY, the gonads develop into ovaries. 11. Describe the inheritance of a sexlinked gene such as color-blindness. Red-Green Color Blindness Pedigree Analysis 12. Explain why a recessive sex-linked gene is always expressed in human males. 13. Explain how an organism compensates for the fact that some individuals have a double dosage of sexlinked genes while others have only one. Mosaicism 14. Distinguish among nondisjunction, aneuploidy, and polyploidy; explain how these major chromosomal changes occur and describe the consequences. Nondisjunction Meiotic Nondisjunction Mitotic Nondisjunction Anaphase I or Anaphase II, One gamete gets 2, the other gets 0 If it occurs in embryonic cells, the abnormal chromosome # may be passed to a large # of cells Anueploidy (trisomy) (monosomy) + = + 0 = An anueploid zygote passes the aberration on to all subsequent cells. A common example is trisomy 21, the cause of Down’s syndrome. Polyploidy 3n = triploidy (fertilization of an egg that had nondisjunction of all chromosomes during oogenesis) 4n = tetraploidy (division of a zygote without cytokinesis) Polyploidy is common in plants, but rare in animals. Polyploid animals are usually mosaics, and more normal in appearance than anueploids. 15. Distinguish between trisomy and triploidy. 16. Distinguish among deletions, duplications, translocations, and inversions. Deletion Duplication Translocation Insertion 17. Describe the effects of alterations in chromosome structure, and explain the role of position effects in altering the phenotype. Alternations of chromosome structure can have various effects: • Homozygous deletions, including a single X in males are usually fatal • Duplications and translocations tend to have deleterious effects • Even if all genes are present in normal dosages, reciprocal translocations between nonhomologous chromosomes can alter the phenotype because of position effects 18. Describe the type of chromosomal alterations implicated in the following human disorders: Down syndrome, Klinefelter syndrome, extra Y, triple-X syndrome, Turner syndrome, cri du chat syndrome, and chronic myelogenous leukemia. Effects of Alterations Down’s syndrome trisomy 21 Kleinfelter syndrome Extra Y XXY, XXXY, XXXXY, XXXXXY XYY Triple X XXX Turner syndrome X0 Cri du chat Deletion on #5 Chronic Myelogenous Leukemia Translocation 22<>9 19. Define genomic imprinting and provide evidence to support this model. It usually doesn’t matter whether a gene is introduced to the next generation via maternal or paternal chromosomes, but for a small number of cases, parental imprinting affects the expression of the gene. A good example of this is the Prader-Willi/Angelman syndromes. 20. Explain how the complex expression of a human genetic disorder, such as fragile-X syndrome, can be influenced by triplet repeats and genomic imprinting. 21. Give some exceptions to the chromosome theory of inheritance, and explain why cytoplasmic genes are not inherited in a Mendelian fashion. The genetic material found in mitochondria and plant plastids, does not follow the pathways of assortment and segregation normal to nuclear chromosomes.