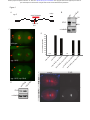
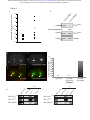
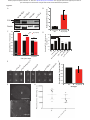
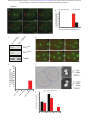
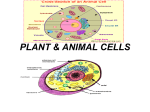
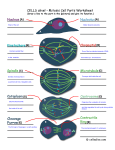
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Protein phosphatase 1 down regulates ZYG
Magnesium transporter wikipedia , lookup
Cellular differentiation wikipedia , lookup
Signal transduction wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Cell growth wikipedia , lookup
Kinetochore wikipedia , lookup
Spindle checkpoint wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Green fluorescent protein wikipedia , lookup
bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Proteinphosphatase1downregulatesZYG-1levelstolimitcentriole duplication NinaPeel1*#,JyotiIyer2#,AnarNaik1,MichaelPDougherty2,MarkusDecker3,KevinFO’Connell2* 1 DepartmentofBiology,TheCollegeofNewJersey,Ewing,NJ,08618. 2 LaboratoryofBiochemistryandGenetics,NationalInstituteofDiabetes,DigestiveandKidney Diseases,NationalInstitutesofHealth,8CenterDrive,Bethesda,MD20814 3 Currentaddress:GenentechInc.,1DNAWay,CancerImmunology,Mailstop231A,SouthSan FranciscoCA94080 # Theseauthorscontributedequallytothiswork. *Correspondingauthors: NinaPeel KevinO’Connell Condensedtitle:PP1inhibitscentrioleduplication Characters:27,054,excludingspaces [email protected] [email protected] 1 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract In humans perturbations of centriole number are associated with tumorigenesis and microcephaly, therefore appropriate regulation of centriole duplication is critical. The C. elegans homolog of Plk4, ZYG-1,isrequiredforcentrioleduplication,butourunderstandingofhowZYG-1levelsareregulated remainsincomplete.WehaveidentifiedthetwoPP1orthologs,GSP-1andGSP-2,andtheirregulators I-2SZY-2 and SDS-22 as key regulators of ZYG-1 protein levels. We find that down-regulation of PP1 activityeitherdirectly,orbymutationofszy-2orsds-22canrescuethelossofcentrioleduplication associated with a zyg-1 hypomorphic allele. Suppression is achieved through an increase in ZYG-1 levels, and our data indicate that PP1 normally regulates ZYG-1 through a post-translational mechanism. While moderate inhibition of PP1 activity can restore centriole duplication to a zyg-1 mutant, strong inhibition of PP1 in a wild-type background leads to centriole amplification via the productionofmorethanonedaughtercentriole.Ourresultsthusdefineanewpathwaythatlimits thenumberofdaughtercentriolesproducedeachcycle. AuthorSummary The centrosomes are responsible for organizing the mitotic spindle a microtubule-based structure thatcenters,thensegregates,thechromosomesduringcelldivision.Whenacelldividesitnormally possesses two centrosomes, allowing it to build a bipolar spindle and accurately segregate the chromosomestotwodaughtercells.Appropriatecontrolofcentrosomenumberisthereforecrucial to maintaining genome stability. Centrosome number is largely controlled by their regulated duplication.Inparticular,theproteinPlk4,whichisessentialforduplication,mustbestrictlylimited as an overabundance leads to excess centrosome duplication. We have identified protein phosphatase1asacriticalregulatoroftheC.elegansPlk4homolog(knownasZYG-1).Whenprotein phosphatase1isdown-regulated,ZYG-1levelsincreaseleadingtocentrosomeamplification.Thusour workidentifiesanovelmechanismthatlimitscentrosomeduplication. 2 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Introduction Inmitoticcellsthecentrosomeservesastheprimarymicrotubule-organizingcenterandconsistsof two centrioles surrounded by a proteinaceous pericentriolar matrix (PCM). During mitosis the centrosomes organize the poles of the spindle, therefore maintaining appropriate centrosome numbers promotes spindle bipolarity and faithful chromosome segregation. Regulated centrosome duplication is the primary mechanism by which centrosome number is controlled, and involves building a new daughter centriole adjacent to each pre-existing mother centriole. Two features of centrioleduplicationmaintainappropriatecentrosomenumbers:first,centrioleduplicationislimited to occurring only once per cell cycle. Second, only a single daughter centriole is assembled in associationwitheachpre-existingmothercentriole. A conserved set of five centriole duplications factors, SPD-2/CEP192, ZYG-1/Plk4, SAS-6, SAS5/STIL/Ana2,arerequiredfordaughtercentrioleassemblyandtheirindividuallossresultsincentriole duplication failure (reviewed in [1]). Conversely, individual over expression of a subset of these duplicationfactors,Plk4,SAS-6andSTIL/SAS-5,leadstocentrioleover-duplication(theproductionof more than a single daughter) leading to a condition known as centriole amplification (the accumulation of an excess number of centrioles) [2-6]. Interestingly, the three factors whose overexpressionleadstocentrioleamplificationhavebeenidentifiedaskeyplayersintheinitialsteps ofcentrioleduplication.InhumancellsPlk4phosphorylatesSTILtotriggercentriolarrecruitmentof SAS-6, which initiates formation of the cartwheel, the central scaffolding structure of the new centriole[7-12].SimilarlyinC.elegansthePlk4homologZYG-1recruitsacomplexofSAS-5andSAS-6 throughdirectphysicalassociationwithSAS-6toinitiatecentrioleduplication[13,14]. Because Plk4 overexpression causes the formation of extra daughter centrioles, Plk4 protein levels must be tightly regulated in vivo. One mechanism that regulates Plk4 levels is its SCF-mediated degradation promoted by autophosphorylation, whereby the active kinase induces its own destruction [15-17]. Degradation of Plk4 homologs in Drosophila, (plk4/Sak) and C. elegans (ZYG-1) are similarly regulated by their SCF-mediated targeting for degradation [18,19]. In addition, recent studies have shed light on temporal and spatial regulation of Plk4 levels. Plk4 initially localizes in a broadringaroundthemothercentrioleuntil,coincidentwiththeinitiationofduplication,itbecomes restricted to a small focus marking the location of daughter centriole assembly [8,20,21]. Emerging 3 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. evidencesuggeststhatthistransition,whichseemstobeakeystepinensuringonlyasingledaughter centriole is assembled, relies on spatially regulated Plk4 degradation [8,22]. Because STIL can both activate Plk4 [10,22,23], and also protect it from degradation [22] it is proposed that centriolar recruitment of a small focus of STIL at the G1/S transition triggers broad Plk4 activation and degradation via autophosphorylation, while protecting a local focus. Thus STIL limits the centriolar distributionofPlk4,promotingtheassemblyofasingledaughtercentriole.Thesestudiesrevealthe central role that regulated destruction of ZYG-1/Plk4 plays in controlling centriole number, and highlighttheimportanceofbetterunderstandinghowthestabilityofPlk4isregulated. Protein phosphatase 1 (PP1) is a major cellular phosphatase that plays well-characterized roles in diverseprocessesincludingglycogenmetabolism,circadianrhythmsandcelldivision.Humanspossess a single PP1α gene, a single PP1β gene (also known as PP1δ) and a single PP1γ gene. The different isoforms are >85% identical and show largely overlapping roles, although some functional specialization has been identified [24]. C. elegans has four PP1 catalytic subunits: a single broadlyexpressed PP1β homolog, GSP-1, and three PP1α homologs, GSP-2, which is also widely expressed, and GSP-3 and GSP-4, whose expression is limited to the male germ line [25,26]. The core catalytic subunitofPP1candirectlybindasubsetofsubstrates,butfunctionalspecificityislargelyconferred byitsinteractionwithregulatoryproteins.Over200PP1interactorsexist,whichregulatePP1through modulating substrate specificity, enzyme localization or by inhibition or activation of phosphatase activity[27].TwoevolutionarilyconservedPP1regulatorsthatplayaroleincelldivisionareinhibitor 2 (I-2) and SDS22. I-2 was originally identified as an inhibitor of PP1 and shows potent inhibitory activityinvitro,althoughinterestinglytheyeasthomologofI-2,GLC8,canalsostimulatePP1activity [28-30].DownregulationofI-2inDrosophilaorhumancellsleadstochromosomemis-segregation, which is proposed to result from mis-regulation of Aurora B [31,32]. Similarly SDS22 antagonizes Aurora B autophosphorylation, downregulating Aurora B kinase activity [33]. Although it has been noted that PP1α regulates centrosome cohesion, [34,35], no role for PP1 in regulating centrosome duplicationhaspreviouslybeenfound. HerewereportanovelPP1-dependentpathwaythatplaysacriticalroleinensuringthateachmother centriole produces one and only one daughter centriole during each cell cycle. We show that PP1 togetherwithtwoofthemostconservedPP1regulators,I-2andSDS-22downregulatesZYG-1protein 4 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. abundance. Our data indicate that PP1 regulates ZYG-1 levels post-translationally and we demonstrate that loss of PP1-mediated regulation leads to ZYG-1 overexpression and centriole amplificationthroughtheproductionofmultipledaughtercentrioles. Results LossofI-2szy-2,aconservedPP1regulator,rescuesthecentrioleassemblydefectofzyg-1(it25)embryos ZYG-1 is essential for centriole assembly: when hermaphrodites carrying the temperature sensitive zyg-1(it25) mutation are grown at the non-permissive temperature of 24°C centriole duplicationfails,resultingin100%embryoniclethality.Toidentifyadditionalregulatorsofcentriole assembly,wescreenedforsuppressorsofthezyg-1(it25)phenotypeandisolatedmutationsin20szy (suppressorofzyg-1)genesthatrescueembryonicsurvival[36].Wemappedoneofthesemutations, szy-2(bs4),totheY32H12A.4locusonchromosomeIII,hereafterreferredtoasszy-2.Theszy-2(bs4) mutationisasinglebasepairchange(GtoA)inasplicedonorsite,whichispredictedtoaltersplicing oftheszy-2transcriptresultinginaframe-shift(Fig1A).AnalysisoftheSZY-2sequenceshowsthatit is homologous to inhibitor-2 (I-2), a conserved regulator of protein phosphatase 1 [30]. Using antibodiesraisedagainstI-2SZY-2weconfirmedthattheszy-2(bs4)mutationsignificantlyreducesI-2SZY2 protein levels, although residual I-2SZY-2 is still detected, indicating that at least some message is properly spliced in the mutant (Fig 1B). At 24°C the embryonic viability of zyg-1(it25); szy-2(bs4) double mutant embryos is 83%, significantly higher than zyg-1(it25) (0% viability). To determine if suppression of the embryonic lethal phenotype was associated with restoration of centriole duplication, we imaged zyg-1(it25) and zyg-1(it25); szy-2(bs4) embryos expressing GFP::tubulin and mCherry::histone(Fig1C).Normallyatfertilizationthespermdeliversasinglepairofcentriolestothe acentrosomalegg,andduringthefirstcellcyclethecentriolesseparate,duplicateandformthepoles of the mitotic spindle (Fig 1C (top panel), Fig S1A and movie S1). Subsequent duplication events ensure the formation of bipolar spindles in later divisions. When the first round of centriole duplication fails, as in zyg-1(it25) mutants, the two sperm-derived centrioles still separate and organize the poles of the first mitotic spindle leading to a normal first division. However as each daughter cell inherits a single centriole, monopolar spindles assemble in the 2-cell embryo (Fig 1C (middlepanel),FigS1BandmovieS2).Asthisphenotypeisindicativeofcentrioleduplicationfailure, we scored the presence of monopolar spindles in 2-cell stage embryos and found that >80% of centrioles duplicated during the first round of duplication in zyg-1(it25);szy-2(bs4) double mutant 5 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. embryos (Figs 1C (bottom panel), 1D & movie S3). In contrast, this first duplication event always failed in zyg-1(it25) control embryos (Fig 1C (middle panel) & D). We confirmed that this effect is specificbyusingRNAitodepleteSZY-2inzyg-1(it25)worms;RNAiofszy-2butnotthenon-essential gene smd-1 (control RNAi), restored centriole duplication to the zyg-1(it25) mutant (Fig 1D). Moreoverthedeletionalleleszy-2(tm3972),inwhichtheC-terminal190residuesareremoved,was also able to suppress the zyg-1(it25) phenotype (Fig 1D). Together these data demonstrate that reducingI-2SZY-2functionsuppresseszyg-1(it25)andsuggestthatmodulatingPP1activitymayimpact centrioleduplication. Figure1.Loss-of-functionmutationsinthePP1regulatorI-2SZY-2rescuesthezyg-1(it25)phenotype. A)Scalediagramofthestructureoftheszy-2gene,indicatingthelocationofthetm3972deletionand theszy-2(bs4)splicesitemutation.B)Westernblotofembryoextractsfromwild-typeandszy-2(bs4) mutants showed a reduced level of the SZY-2 protein. C) Stills from movies of embryos expressing mCherry::H2BandGFP::tubulingrownat24°C.Top,wildtype;Middle,zyg-1(it25)mutantatthe2-cell stagefollowingcentrioleduplicationfailure;bottom,zyg-1(it25);szy-2(bs4)doublemutantsat2cellstageshowingarescueofthecentrioleduplication.D)Quantificationofcentrioleduplicationfailure at 24°C in zyg-1(it25) mutants and in zyg-1(it25) mutants in which szy-2 activity has been downregulated by mutation or RNAi. Number of centriole duplication events analyzed is indicated. E) Western blot of embryo extracts, showing levels of phospho-histone H3 in wild type and szy-2(bs4) embryos.F)Phospho-histoneH3stainingofwild-typeandszy-2(bs4)embryosatfirstmetaphase.Left, merge:DNAblue,Microtubulesred,Phospho-histoneH3green.Right,phospho-histoneH3. Lossofszy-2decreasesPP1activity Because szy-2 encodes the C. elegans homolog of I-2, this suggested that the szy-2(bs4) allele may alter PP1 activity, resulting in the observed suppression of centriole duplication failure. I-2 is conservedfromyeastthroughhumansandhasbeendescribedasbothaninhibitorandanactivator ofPP1[28-30].AnalysesinvitrohaveshownthatC.elegansI-2SZY-2caninhibitrabbitPP1activity,but theinvivoroleofszy-2,inparticularinrelationtoendogenouswormPP1subunits,hasnotpreviously been investigated [30]. We sought to determine whether the important function of I-2SZY-2 with respecttozyg-1(it25)suppressionwasasaninhibitororanactivatorofPP1.Todeterminewhether PP1 activity is up- or down-regulated in the szy-2(bs4) mutants we wanted to monitor the 6 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. phosphorylation levels of a known PP1 substrate. One such substrate is histone H3, which is phosphorylated in the early stages of mitosis; this modification is removed by PP1 [37,38]. To investigate whether I-2SZY-2is an activator or inhibitor of PP1 we analyzed phospho-histone levels in szy-2(bs4) mutant embryos. We used a phospho-histone H3 antibody to detect phosphorylated histone H3 levels in embryos and found them to be greatly elevated in szy-2(bs4) when compared with the wild type (Fig 1E & F), a result reminiscent of PP1 depletion (Fig 3A; [25,39]). Phosphohistonelevelswereelevatedinbothmixedstageembryos(Fig1E)aswellasduringthemetaphase stageofmitosis(Fig.1F).Thisresultindicatesthattheszy-2(bs4)mutationreducesPP1activityand suggests that I-2SZY-2 normally acts as an activator of PP1. Consistent with I-2SZY-2 being a positive regulatorofPP1,szy-2(bs4)mutantembryosexhibitachromosomemis-segregationdefect[36]thatis similartothatofembryosdepletedofPP1(movieS4).ThereisprecedentforI-2actinganactivatorof PP1:inyeastI-2GLC8bindsandinactivatesPP1GLC7,butuponphosphorylationofI-2GLC8,PP1becomes activated [29]. The phosphorylated residue is conserved in all I-2 homologs including C. elegans, althoughitisnotclearwhetherI-2SZY-2issimilarlyregulatedintheworm[30]. LossofasecondPP1regulator,sds-22,rescuesthecentrioleduplicationfailureofzyg-1(it25)embryos BecausewehadidentifiedamutationinI-2szy-2asasuppressorofzyg-1(it25),wespeculatedthatloss ofadditionalPP1regulatoryproteinsmayhaveasimilareffect.Totestthishypothesisweidentified sevenC.elegansproteinswhichshowahighdegreeofconservationwithhumanPP1regulators.We thenusedRNAitodepleteeachfactorinzyg-1(it25)worms,andmonitoredembryonicviability(Fig S2). RNAi of only the sds-22 gene significantly increased embryonic viability of the zyg-1(it25) embryos. SDS-22 is a leucine-rich repeat (LRR) domain containing protein that localizes PP1 during mitosis,regulatingmitoticprogression[33,40-42].Althoughsds-22(RNAi)hadonlyamodesteffecton zyg-1(it25)embryonicviability,directobservationofcentrioleduplicationinzyg-1(it25);sds-22(RNAi) embryos, revealed 70% of centriole duplication events occurred normally (Fig 2B). The disparity between the strength of suppression of embryonic lethality and the strength of suppression of centrioleduplicationfailureislikelyexplainedbyacombinationoftwofactors.First,wefindthaton average,30%oftheduplicationeventsinzyg-1(it25);sds-22(RNAi)embryosfail;suchahighdegreeof celldivisionfailureduringearlydevelopmentwouldresultinembryoniclethality.Second,sincesds-22 is an essential gene, RNAi knockdown may cause embryonic lethality even though centriole duplication is normal. Since reducing SDS-22 function is able to suppress the zyg-1(it25) phenotype 7 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. wehypothesizedthatoneofthesuppressorsofzyg-1(it25)recoveredinourgeneticscreen[36]may have a mutation in sds-22. We therefore sequenced the sds-22 coding region in those szy mutants that mapped close to the sds-22 genetic locus. The szy-6(bs9) strain contained a G-to-A missense mutation in the coding region of sds-22 which results in a single amino-acid substitution (G224S) within the eighth LRR (Fig 2A). To confirm that bs9 was an allele of sds-22 we performed a complementation test with sds-22(tm5187) a deletion allele that exhibits a fully-penetrant larvallethal phenotype (Figs 2A & C). While the szy-6(bs9) homozygotes exhibited minimal embryonic lethality,thetrans-heterozygotes(bs9/tm5187)exhibited100percentembryoniclethalityconfirming thatbs9andtm5187areallelic(Fig2C).Wehavethereforerenamedtheszy-6(bs9)allelesds-22(bs9). Although by itself, the sds-22(bs9) mutation does not appear to affect centriole duplication, zyg1(it25); sds-22(bs9) double mutant embryos exhibit an approximately 50% success rate of centriole duplication, comparable to levels seen after SDS-22 RNAi depletion (Fig 2B). Together these data showthatreducingthefunctionofeitheroftwoPP1regulatoryproteins,I-2SZY-2orSDS-22,isableto partiallysuppressthefailureofcentrioleduplicationcausedbythezyg-1(it25)mutation. Figure 2. Reducing SDS-22 activity rescues the zyg-1(it25) phenotype. A) Scale diagram of the structure of the sds-22 gene, indicating the location of the tm5187 deletion and the sds-22(bs9) substitution.B)Quantificationofcentrioleduplicationwhenzyg-1(it25)isrescuedbythesds-22(bs9) mutation or sds-22(RNAi). Number of centriole duplication events analyzed is indicated. C) Complementation test showing bs9 and tm5187 are allelic. Hermaphrodites of the indicated genotypewereshiftedto25°CattheL4stageandembryoniclethalitywasdeterminedoverthenext 24hours.Theszy-6(bs9)allelewasmarkedwiththecloselylinkeddpy-10(e128)mutation. LossofPP1βGSP-1activityrescueszyg-1(it25) OurdatasuggestthatI-2SZY-2isanactivatorofPP1,andworkinvertebratecellshasshownthatthe SDS-22 ortholog positively regulates PP1 activity [33]. Because the szy-2(bs4) and sds-22(bs9) mutations rescue zyg-1(it25) phenotypes it followed that directly lowering PP1 activity should also suppress the zyg-1(it25) phenotype. Embryos express two PP1 catalytic subunits encoded by the genesgsp-1andgsp-2.Similartowhatisseenintheszy-2(bs4)mutant(Fig1E&F),co-depletionof GSP-1 and GSP-2 by RNAi resulted in an increase in phospho-histone H3 staining of mitotic chromosomes(Fig3A).WenexttestedwhetherindividuallyreducingtheactivityofeitherPP1GSP-1or 8 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. PP1GSP-2, could suppress the centriole duplication failure seen in zyg1(it25) embryos. Although the gsp-1(tm4378)deletionalleleissterile,precludingitsuseinouranalysis,wewereabletospecifically depletePP1GSP-1byRNAi(Fig3B).WhenPP1GSP-1wasdepletedinzyg-1(it25)embryoswesawarobust restorationofcentrioleduplication(90%ofcentrioles;Fig3B,C,D&G).Incontrast,theputativenull allelegsp-2(tm301)[25]didnotsuppressthezyg-1(it25)phenotypeandweneverobservedcentriole duplication in the zyg-1(it25); gsp-2(tm301) double mutant embryos (Fig 3E, F & G). The C. elegans PP1αGSP-2andPP1βGSP-1isoformsshowahighdegreeofidentityandarebothexpressedintheearly embryo [25]. Since depletion of only the PP1βGSP-1 isoform can suppress the zyg-1(it25) allele this suggestseither1)adivergenceinfunctionofthetwoenzymes,withonlyPP1βGSP-1beinginvolvedin the regulation of centriole duplication, or 2) that, despite partial redundancy between the two enzymes, zyg-1(it25) mutants provides a sensitized background where loss of PP1βGSP-1 alone is sufficienttosubvertnormalcontrols.Indeed,twoobservationssupportthelaterpossibility.First,we have only observed defects in chromosome segregation after co-depletion of both PP1βGSP-1 and PP1αGSP-2 suggestive of some functional redundancy (movie S4). Second, we show below that codepletion of PP1βGSP-1and PP1αGSP-2 disrupts the normal pattern of centriole duplication whereas singledepletionsofeitherphosphatasehavenoeffect(Fig5C,D,E&movieS5). Figure 3 PP1 functions with I-2SZY-2and SDS-22 to regulate centriole duplication. A)Comparisonof phospho-HH3 levels in control and PP1βGSP-1 & PP1αGSP-2 co-depleted embryos as determined by quantitative immunofluorescence microscopy. The mean is indicated by the bar. B) Western blot demonstrating specificity of gsp-1(RNAi). C&D) Stills from time-lapse recordings of embryos of the indicated genotypes, expressing GFP::histone and GFP::SPD-2 grown at the normally restrictive temperatureof24°C.E&F)Stillsfromtime-lapserecordingsofembryosoftheindicatedgenotypes, expressingmCherry::histoneandGFP::tubulingrownatthenormallyrestrictivetemperatureof24°C; centrosomes are indicated by arrow heads. G) Quantitation of centriole duplication in indicated strains. Number of centriole duplication events scored is indicated. H) Western blots of immunoprecipitated material from whole worm extracts using control IgG, I-2SZY-2, or PP1βGSP-1 antibodies,andprobedwithI-2SZY-2,PP1βGSP-1,orSDS-22antibodiesasindicated.I)Westernblotsof immunoprecipitated material from whole worm extracts using control IgG, I-2SZY-2, or SDS-22 antibodies,andprobedwithI-2SZY-2orPP1αGSP-2,orSDS-22antibodies. 9 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Our data indicate that individually reducing the function of either I-2SZY-2, SDS-22 or PP1βGSP-1 can partially suppress the zyg-1(it25) centrosome duplication failure. Using GFP fusions we were, however, unable to detect centrosome localization of any of the proteins (Fig S3). Nevertheless we reasoned that all three proteins work in a common process and sought to determine whether they physically interact. When we immunoprecipitated I-2SZY-2 from C. elegans embryonic extracts, we found by immunoblotting that we could detect co-precipitated PP1βGSP-1 and reciprocally when we immunoprecipitatedPP1βGSP-1wewereabletopulldownI-2SZY-2(Fig3H),confirmingthatC.elegansI2SZY-2isaPP1bindingproteininvivo.Similarly,antibodiesagainstPP1βGSP-1co-precipitatedSDS-22(Fig 3H).However,wewereunabletodetectanyinteractionbetweenI-2SZY-2andSDS-22,suggestingthat a complex simultaneously containing all three proteins does not form. Additional IP-western experimentsdemonstratedthatI-2SZY-2alsointeractswithPP1αGSP-2,butdidnotrevealaninteraction between SDS-22 and PP1αGSP-2 (Fig 3I). To confirm and extend these results we also individually immunoprecipitatedI-2SZY-2,SDS-22,andPP1βGSP-1andanalyzedtheimmunoprecipitatedmaterialby massspectrometry(FigS4).UsingthisapproachwefoundthatI-2SZY-2andSDS-22couldindependently interact with both PP1βGSP-1 and PP1αGSP-2 but not with each other (Fig S4). We conclude that componentsofthePP1-dependentregulatorypathwayphysicallyinteractbutthatI-2SZY-2andSDS-22 donotappeartoresideinthesamecomplex. Finally,wehavefoundthatalthoughbothI-2SZY-2andSDS-22interactwithPP1andpositively regulatePP1’sfunctionincontrollingcentrioleduplication,thetworegulatorsdonotalwaysfunction togethertocontrolPP1activity.SpecificallywehavefoundthatwhilelossofI-2SZY-2activityresultsin anincreaseinthelevelofphospho-histoneH3inmitoticchromosomes(Fig1F),nosuchincreaseis observeduponlossofSDS-22activity(FigS5B).ThusI-2SZY-2appearstofunctionindependentlyofSDS22 to mediate the role of PP1 in regulating histone H3 phosphorylation. Curiously, loss of SDS-22 resultedinanelevatedphospho-histoneH3signalasdetectedbyimmunoblotting(FigS5A).Because SDS-22doesnotcontrolthechromatincontentofphospho-histoneH3,wespeculatethatlossofSDS22 leads to a mitotic delay; this would explain the elevated signal of phospho-histone H3 in mixedstagewormextracts. AuroraB,AuroraAandPlk1arenotrequiredforszy-2(bs4)suppressionofzyg-1(it25) 10 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. GiventhatPP1isamitoticphosphatasethatantagonizesmanyknownmitotickinaseswewantedto investigate whether these relationships contribute to the regulation of centriole duplication. In Drosophila and human cells, I-2 is implicated in regulation of Aurora B and its depletion results in chromosome mis-segregation [31,32]. Similarly in humans, SDS22 antagonizes Aurora B autophosphorylation, downregulating Aurora B kinase activity [33]. PP1, I-2 and SDS22 therefore seem to cooperatively regulate chromosome segregation by modulating Aurora B activity. In C. elegans PP1 also regulates Aurora B, contributing to its correct localization during meiosis and to chromosomesegregationinmitosis[39,43].AlthoughAuroraBhasnotbeenimplicatedincentriole duplication we wanted to determine whether szy-2(bs4) suppresses centriole duplication failure throughupregulationofAuroraBactivity.WethereforeRNAi-depletedAuroraBAIR-2inszy-2(bs4);zyg1(it25)wormsandmonitoredcentrioleduplication(FigS6A,B&C).Althoughweobservedfailuresin chromosomesegregationandcytokinesisconsistentwithsuccessfulAuroraBdepletion(FigS6C;[44]), centrioleduplicationwasunperturbed(FigS6A),indicatingthatszy-2(bs4)doesnotregulatecentriole duplicationbymodulatingAuroraBactivity. Since PP1 and I-2 are associated with regulation of Aurora A activity [45] and Aurora A plays a conserved role in centrosome separation and maturation [46,47] we tested whether Aurora AAIR-1 activityisrequiredforsuppressionofzyg-1(it25)byszy-2(bs4).Afterexposingdoublemutantstoair1(RNAi) we observed reduced SPD-2 localization and incomplete centrosome separation, consistent withlossofAuroraAactivity(FigS7H),howeverwedidnotseeaperturbationofcentrioleduplication (Fig S6A, Fa & Fb), suggesting that Aurora A is not required for suppression of zyg-1(it25) by szy2(bs4).InvertebratecellsPP1alsohasarecognizedroleinantagonizingPlk1[48],akinasewithan established role in centriole duplication [49]. However, depletion of plk-1 in zyg-1(it25), szy-2(bs4) embryos did not affect centriole duplication (Fig S6A) even though PLK-1 activity was clearly compromised (Fig S6D). In summary our data suggest that reducing PP1 activity does not restore centriole duplication in the zyg-1(it25) mutant by relieving antagonism, and thus increasing the relativeactivity,ofAuroraB,Plk1orAuroraA,whichareknownPP1antagonists. I-2SZY-2controlsSPD-2levelsatthecentrosome During the course of our analyses we noted that the level of the coiled-coil protein SPD-2 was elevatedatthecentrosomeinszy-2(bs4)mutantembryos.SPD-2isacomponentofboththePCMand 11 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. centrioles and is required for centriole duplication and PCM assembly [50,51]. We quantified the effectonSPD-2levelsinszy-2(bs4)embryosexpressingSPD-2::GFPandfounda1.5-foldincreasein centrosomal SPD-2 levels throughout the cell cycle (Fig S7A). When we compared levels of endogenousSPD-2atthecentrosome,wefoundasimilarincreaseincentrosome-associatedSPD-2in szy-2(bs4)embryos(FigS7B-D).SinceSPD-2playsapositiveroleincentrioleduplication,wereasoned that its increased levels at the centrosome in the szy-2(bs4) mutant might be responsible for PP1mediated suppression. To test this possibility, we utilized a codon-optimized spd-2::gfp transgene [52]tooverexpressSPD-2proteininthezyg-1(it25)mutant.Despitealargeincreaseincentrosomelocalized SPD-2 (Fig S7E & F) we did not see any suppression of zyg-1(it25) embryonic lethality (Fig S7G),indicatingthatcentrioleduplicationfailurepersisted.Thusbyitself,ageneralelevationofthe levelofSPD-2atthecentrosomeisnotsufficientforsuppressionofzyg-1(it25).Conversely,wealso findthattheelevatedlevelofcentrosome-associatedSPD-2isnotrequiredforszy-2(bs4)-mediated suppression:AuroraAAIR-1isrequiredforSPD-2recruitmenttothecentrosome[50],anddepletionof auroraAAIR-1inthezyg-1(it25);szy-2(bs4)doublemutantdrasticallyreducedtheamountofSPD-2at the centrosome, (Fig S7H) nevertheless, centriole duplication was not perturbed (Fig S6A, E & F). Thus,I-2SZY-2regulatesbothcentrioleduplicationandPCMassembly,buttheelevatedSPD-2levelsat thecentrosomeobservedinszy-2(bs4)mutantsdonotconstitutetheprimarymechanismbywhich szy-2(bs4)suppressesthecentrioleduplicationdefectofzyg-1(it25)mutants. ReducingPP1activityelevatesZYG-1proteinlevels Previous work has shown that the failure of centriole duplication in zyg-1(it25) mutants can be rescuedbyincreasingcentriole-associatedlevelsofthemutantZYG-1protein[19,53].Wetherefore sought to determine if decreasing PP1 activity affects ZYG-1 protein levels or localization. To determinewhethertheszy-2(bs4)mutationaffectsZYG-1abundance,wemeasuredtotalZYG-1levels inearlymixed-stageembryosusingquantitativeimmunoblotting.Strikingly,incomparisontothewild type,szy-2(bs4)embryosconsistentlypossessedapproximatelyfour-foldmoreZYG-1attherestrictive temperature of 25°C (Fig 4A & B). We then used quantitative immunofluorescence to see if the elevatedleveloftotalZYG-1proteinresultedinincreasedlevelsofZYG-1atcentrioles.Asexpected, theoverallincreaseinZYG-1levelsinembryoswasassociatedwithelevatedZYG-1atthecentrioles (Fig 4C, D & E). This increase however was cell cycle stage specific with increases observed in prophase and metaphase but not anaphase, of the first cell cycle (Fig 4C). We further sought to 12 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. determine whether decreasing SDS-22 or PP1 similarly increased ZYG-1 levels. Quantification of centrosome-associated ZYG-1 in prophase revealed elevated levels of ZYG-1 in the sds22(bs9/tm5187)trans-heterozygousmutantbutaminimalincreaseafterRNAi-depletionofPP1βGSP-1 (Fig4D&E).SimilarlydepletionofPP1αGSP-2hadlittleeffectonZYG-1levelsatcentrioles(Fig4D&E). Since we only observed chromosome segregation defects when we depleted both PP1βGSP-1 and PP1αGSP-2 (movie S4), we wondered whether they also acted redundantly to regulate ZYG-1. We thereforeco-depletedPP1βGSP-1andPP1αGSP-2andfoundanincreaseincentrosome-associatedZYG-1, consistentwiththeexistenceofredundancybetweenthetwoPP1isoforms(Fig4D&E).Overallour results suggest that reducing PP1 activity leads to an elevation of both total ZYG-1 levels, and of centriole-associated ZYG-1, providing a likely mechanism for the suppression of the zyg-1(it25) centrioleduplicationdefect.Notably,thiseffect,ofreducedPP1activityleadingtoincreasedZYG-1 levels,seemstobespecificaswedidnotseeasimilarincreaseinSAS-6levelsinszy-2(bs4)embryos (FigS8A,B&C).Finally,becauseZYG-1hasalsobeenimplicatedinpositivelyregulatingcentrosome size[53],elevatedZYG-1levelsmayalsoaccountfortheobservedincreaseincentrosomalSPD-2. Figure 4. ZYG-1 protein levels are elevated in the szy-2(bs4) mutant. A) Western blot of embryo extracts from wild type and szy-2(bs4) embryos probed with a ZYG-1 antibody. The ZYG-1 band (arrow)isidentifiedbyitsabsenceinzyg-1(RNAi)extracts.Non-specificproteins(arrowhead)arealso bound by the ZYG-1 antibody and probably represent contaminants from the E. coli strain used for RNAi feeding experiments. B) Normalized ZYG-1 protein levels from two independent experiments. Errorbarsindicatestandarddeviation.C)Relativelevelsofcentriole-localizedZYG-1inwild-typeand szy-2(bs4) one-cell embryos made by quantitative immunostaining. Levels at each stage are normalized to the wild-type control. Number of centrosomes analyzed indicated inside each bar. **p<0.01; n.s. no significant difference (Student’s t-test). D) Centriole-localized ZYG-1 protein levels for embryos of indicated genotypes at first mitotic prophase. Levels normalized to the wild-type control. Number of centrosomes analyzed indicated inside each bar. **p<0.01; n.s. no significant difference from WT (Student’s t-test). E) Representative images of centrosomal ZYG-1 levels in prophase.Centrosomesfrom1-cellstageembryosoftheindicatedgenotypesaregroupedwithawild typesamplefromthesameexperiment.F)Quantificationofzyg-1transcriptlevelsinembryosbyqRTPCR.Shownistheaverageoftwoindependentexperiments.Errorbarsindicatestandarddeviation. G)Representativeimagesofwild-typeandszy-2(tm3972)embryosexpressingGFP::histonedrivenby 13 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. thezyg-1promoterand3’-UTR.H)QuantitationofGFPintensitieswithmeanandstandarddeviations indicated.Nodifferencebetweenstainswasfound(Student’st-testp>0.1). PP1regulatesZYG-1levelspost-translationally We next sought to determine how ZYG-1 levels are regulated by PP1. First, we measured zyg-1 transcriptlevelsbyquantitativeRT-PCRandfoundthatwild-typeandszy-2(bs4)embryospossessed similar levels of zyg-1 mRNA (Fig 4F). The observed increase in ZYG-1 protein levels was not, therefore,duetoaneffectontranscriptionormRNAstability.TotestwhetherdecreasingPP1activity resultedinaneffectonZYG-1translationefficiency,wecrossedtheszy-2(tm3972)alleleintoastrain carrying a GFP::histone reporter expressed under the control of the zyg-1 regulatory sequences (promoterandUTRs)[54].Sincethisreportercontainszyg-1regulatorysequences,butlacksthezyg-1 coding sequence, it allows us to determine whether elevated ZYG-1 protein levels stem from alterations at the level of gene expression (transcription/translation) or from post-translational controls. Quantitative fluorescence intensity measurements of GFP::histone revealed that the szy2(tm3972)mutationdidnotincreaseexpressionofthereporterrelativetothatobservedinthewildtypestrain(Fig4G&H;Student’st-testp>0.1),indicatingthatreducedPP1activitydoesnotincrease zyg-1 translation via the 3ʹ-UTR. In order to determine whether PP1 might directly regulate ZYG-1 levelswetestedforaninteractionbetweenthetwoproteins.ImmunoprecipitationofI-2SZY-2,SDS-22, and PP1βGSP-1 from worm extracts, followed by western blotting or mass spectrometry failed to detectaninteractionbetweenanyofthethreeproteinsandZYG-1.BecauseZYG-1isalowabundance protein [14] and interactions between PP1 and its substrates may be transient we cannot however ruleoutadirectinteractionbetweenZYG-1andPP1.Cumulatively,therefore,ourdatapointtoPP1 regulatingZYG-1proteinlevelseitherdirectlyorindirectlyviaapost-translationalmechanism. ReductionofPP1activitycausescentrioleamplification Elevated levels of the human and Drosophila homologs of ZYG-1 are associated with centriole amplification[2,5,6].AlthoughcentrioleamplificationhasnotpreviouslybeenobservedinC.elegans embryos,giventheincreasesinZYG-1levelsweobservedintheszy-2(bs4)andsds-22(bs9/tm5187) mutants we were intrigued whether this would be sufficient to subvert the normal regulation of centriole duplication, resulting in supernumerary centrosomes. We therefore performed time-lapse confocalmicroscopyofembryosexpressingGFP::SPD-2andmCherry::histone.Initialinspectionofszy 14 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. 2 and sds-22 mutants did not reveal any obvious defects during the first two cell cycles; centriole duplicationproceedednormallyandbipolarspindlesassembledinallcells.Unexpectedlyhowever,in embryos strongly impaired for either I-2SZY-2or SDS-22 function, extra centrosomes were frequently observedatthefour-cellstage(Fig5A,B&movieS6).Closerinspectionofthemoviesindicatedthat the extra centrosomes arose approximately synchronously from the spindle poles as the PCM dispersednearthecompletionofthesecondmitoticdivisions(moviesS6&S7).Inasinglecase(out of79analyzed)wewerenotabletotracetheoriginofoneofthesecentrosomestoaspindlepole. Thus, it is possible that very infrequently in this mutant a centriole arises spontaneously in the cytoplasm via a de novo pathway. The production of extra centrosomes was most prevalent in sds22(bs9/tm5187) trans-heterozygotes where nearly 40% of the spindle poles gave rise to more than twocentrosomes(Fig5B).Althoughlessfrequent,anidenticaldefectwasobservedinszy-2(tm3972) embryoswheresupernumerarycentrosomesarosewiththesametimingandspatialpatternasthose observed in the sds-22 mutant (Fig 5B). These results suggest that extra centrioles form during the second round of centriole duplication (which occurs in the 2-cell embryo) such that the extra centrioles only become apparent by confocal microscopy when mother and daughter centrioles separateascellsenterthethirdcellcycle(FigS1C). Wenextfollowedthefateof79centrosomesinsds-22(bs9/tm5187)trans-heterozygotesandfound that all eventually accumulated PCM and participated in spindle assembly, often giving rise to multipolar spindles (movie S7). A minority (4/79 centrosomes), however, exhibited a one-cell-cycle delay before accumulating normal levels of PCM and participating in spindle assembly (Fig S9 and movie S7). This is somewhat reminiscent of the reversible “inactivation” of centrioles observed in Drosophila embryos following over-expression of Plk-4 [55]. The production of excess centrosomes continued in the ensuing cell cycles. We quantified the rate of over-duplication following the third centriole cycle of sds-22(bs9/tm5187) embryos and found a similar frequency of over-duplication (35%, n= 96 centrosomes). Thus our results suggest that over-duplication begins during the second centrioleduplicationeventandcontinuesthroughembryonicdevelopment. Toconfirmthattheexcesscentrosomesobservedinszy-2andsds-22mutantembryosaroseduetoa reductionofPP1activity,wealsofollowedcentrosomesduringthefirstseveralcellcyclesofembryos depleted for one or both PP1 catalytic subunits. Surprisingly, neither depletion of PP1βGSP-1 (n=10 15 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. embryos) nor of PP1αGSP-2 (n=11 embryos) resulted in the appearance of extra centrosomes during the first three cell cycles (Fig 5E). We therefore co-depleted both catalytic subunits (Fig. 5C) and found that excess centrosomes appeared at the four-cell stage in 4/10 embryos (Fig 5D, E & movie S5).Further,theseextracentrosomescouldfunctionasmicrotubule-organizingcentersanddirected the formation of tripolar spindles (movie S8). Interestingly, we also observed occasional anaphase chromatin bridges in these embryos (movie S4), whereas in singly depleted embryos chromosomes always segregated normally. We conclude that PP1βGSP-1 and PP1αGSP-2 exhibit some level of functionalredundancyintheirrolesinchromosomesegregationandcentrioleduplication. Figure 5. Decreasing PP1 activity leads to centriole overduplication. A) Frames from a time-lapse recording of an sds-22(bs9/tm5187) trans-heterozygous embryo expressing GFP::SPD-2 and mCherry::histone. Supernumerary centrosomes appear at the 4-cell stage. B) Quantification of centrosome over-duplication in sds-22(bs9/tm5187) and szy-2(3972) embryos during the first and secondroundsofcentrioleduplication.Forsds-22(bs9/tm5187),n=31/77(firstround/secondround) and for szy-2(3972), n= 42/68 (first round/second round) C) Immunoblot showing extent of knockdownofPP1βGSP-1andPP1αGSP-2ingsp-1;gsp-2doubleRNAiembryos.D)Selectedframesfrom a time-lapse recording of a gsp-1(RNAi); gsp-2(RNAi) embryo expressing GFP::histone and mCherry::SPD-2. Arrowheads indicate extra centrosomes. E) Quantitation of the frequency of supernumerarycentriolesinembryosdepletedoftheindicatedPP1genes.F)Structuredillumination microscopy (SIM) of a late 2-cell stage sds-22(bs9/tm5187) trans-heterozygous embryo reveals the presence of excess centrioles. The low magnification image on the left shows the positions of the centrioles stained for the centriole marker SAS-4. Images on the right are enlargements of the indicatedcentrosomes.G)QuantificationofSIMdatatoindicatethenumberofcentriolesobserved percentrosomecomparingwildtypeandsds-22(bs9/tm5187)transheterozygousmutants. Tofurtherinvestigatetheoriginoftheexcesscentrosomesweanalyzedsds-22(bs9/tm5187)embryos by structured illumination microscopy (SIM), which has proven an effective means to observe centriole arrangement and number within the centrosome [56,57]. SIM imaging of SAS-4-stained embryosallowedustoresolvethebasicstructureofwormcentriolesandwecouldclearlydetectthe normal arrangement of mother and daughter centrioles (Fig 5Fa). Strikingly, in sds-22(bs9/tm5187) embryoswewerealsoabletofindmothercentriolesbearingmorethanonedaughter(Fig5Fb).We 16 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. first detected extra daughter centrioles at the late 2-cell stage, consistent with the appearance of excesscentrosomesatthefour-cellstage.Furthermore,theappearanceofextradaughtercentrioles in association with a single mother is reminiscent of what is seen after Plk4 overexpression [2] and indicatesthattheexcesscentrosomesweobserveinthe4-cellembryooriginatefromtheformation ofextradaughtersduringcentrioleduplication.QuantificationofourSIMdatarevealsthatwhilewildtypeembryosneverexceedtwocentriolespercentrosome,18%ofsds-22(bs9/tm5187)centrosomes contain more than 2 centrioles (Fig 5G). We did not observe any other unusual centriole configurations such as mother-daughter-granddaughter arrangements indicative of reduplication of centriolesduringasinglecellcycle.Insummary,ourdataindicatethatlossofPP1activityresultsin overexpression of ZYG-1 and consequently centriole amplification. Amplification appears to be largely,ifnotentirely,drivenbytheproductionofmultipledaughtercentrioles.However,wecannot ruleoutthatcentriolereduplicationanddenovoformationalsocontributetotheexcesscentrioles observed after PP1 inhibition. In conclusion, we have described a new PP1-dependent mechanism that limits centriole duplication so that each mother produces one and only one daughter per cell cycle. Discussion WehaveshownthatPP1activityisanimportantregulatorofZYG-1levelsintheC.elegansembryo andthereforethatitisacriticalregulatorofcentrioleduplication.AlthoughPP1αhaspreviouslybeen implicatedintheregulationofcentrosomeseparationatthebeginningofmitosis[34],thisisthefirst indication that PP1 regulates centriole duplication. Our data indicate that PP1 inhibits centriole duplicationbyrestrainingtheaccumulationofZYG-1intheembryoeitherdirectlyorindirectly(Fig6). PreviousworkhasshownthatPlk4levelscanberegulatedbySCF-mediateddegradationpromotedby autophosphorylation[15-17].SimilarlyinC.eleganstheSCFcomplexisinvolvedinregulatingZYG-1 levels and depletion of SCF components leads to an increase in centrosome-associated ZYG-1 [19]. Ourfindingthatdown-regulationofPP1activityleadstocentrosomeamplificationimplicatesPP1as anadditionalregulatorofZYG-1levels. Figure6.ModelofhowI-2SZY-2,SDS-22andPP1mightcooperatewiththeSCFtoregulatecentriole duplication. Our work shows that PP1, I-2SZY-2 and SDS-22 down regulate ZYG-1 levels to constrain centriole duplication. We speculate that this is a new mechanism of regulation that operates 17 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. independentlyofthepreviouslydocumentedSCF-mediatedregulation.Wecannotruleout,however, aninterplaybetweenthetwopathways(dottedline). How does PP1 regulate ZYG-1 levels? Although many PP1 targets have been identified none are known regulators of centrosome duplication. A major substrate of PP1, I-2 and SDS22 is Aurora B kinaseandactivityoftheC.eleganshomolog,AIR-2,isinhibitedbyPP1[25,39,43].Downregulation ofAIR-2activityinthezyg-1(it25);szy-2(bs4)doublemutanthoweverdidnothaveanymeasurable effectonthelevelofcentrioleduplicationsuggestingthatPP1isnotregulatingcentrioleduplication byantagonizingAuroraBactivity.Identicalresultswereobtainedfromtwoadditionalmitotickinases, Plk1 or Aurora A. Our data indicate that PP1 regulates ZYG-1 levels through a post-translational mechanism.IntheoryPP1couldregulatetherecognitionofZYG-1bytheSCF,howeverwethinkthat thisisunlikelyasourresultsdonotmatchwiththecanonicalmechanismforSCFregulation:substrate recognitionbytheSCFisregulatedbyphosphorylation,buttheincreasedphosphorylationassociated with PP1 down-regulation would be expected to increase proteosomal degradation, decreasing substrateaccumulation.We,however,findanincreaseinZYG-1levelswhenPP1activityisreduced. Furthermore, evidence in Drosophila and C. elegans indicates that degradation of Plk4/ZYG-1 is antagonizedbyPP2A[58,59].Therefore,ourfavoredhypothesisisthatPP1regulatesZYG-1protein levels through an SCF-independent mechanism, perhaps by removing a stabilizing phosphorylation, however the identity of the opposing kinase remains unknown. We cannot, however, rule out the possibilitythatPP1functionsthroughtheSCFpathwayinanon-canonicalfashiontomediateZYG-1 degradation.Ofnote,aKVXFconsensussiteforPP1binding[60]doesexistinZYG-1,howeveritisnot conservedevenincloselyrelatednematodespecies.Nevertheless,althoughthisisacommonmotif bywhichPP1interactswithitssubstratesthepresenceofthismotifisnotanabsoluterequirement forPP1binding. OurdatasuggestthatPP1,I-2SZY-2andSDS-22cooperatetoregulateoverallcellularlevelsofZYG-1. DownregulationofPP1activityleadstoaccumulationofexcessZYG-1andinembryosexpressinga wild-typeversionofZYG-1,theelevatedactivityissufficienttodrivecentrioleover-duplication.Thisis similartothesituationinhumansandDrosophilawhereoverexpressionofhomologsofZYG-1causes centriole over-duplication [2,5,6]. In agreement with this previous work [2] the appearance of supernumerarycentrosomesinC.elegansresultsfromtheformationofextradaughtercentriolesin 18 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. associationwithasinglemother.Intriguingly,wefindthatwhenPP1activityisdecreased,theinitial duplication event (that occurs during the first cell cycle following the female meiotic division) is unaffected, yielding bipolar spindles at the two-cell stage. However, beginning with the second centrioleduplicationevent(thatoccursatthetwocellstage),centriolescommenceover-duplicating resultinginmultipolarspindles,abnormaldivisions,andultimatelylethality. WhyisthefirstduplicationeventunaffectedbythelossofPP1-mediatedregulationwhiletheensuing duplication events go awry? One unique feature of the first duplication event is that it involves paternally-inherited(sperm)centrioleswhilelatereventsinvolvecentriolesassembledintheembryo. Thesperm-derivedcentriolesarenot,however,permanentlyimmunetoelevatedZYG-1levels,aswe have observed cases where, during the second centriole cycle, all four mother centrioles (the two originalsperm-derivedcentriolesplusthetwocentriolesproducedduringthefirstduplicationevent) produce multiple daughters. Thus, it seems that the elevated level of ZYG-1 present in the embryo onlytriggersover-duplicationofthesperm-derivedcentriolesfollowingaone-cell-cycledelay.There are at least three possibilities to explain the pattern of over-duplication in PP1-compromised embryos. First, loss of PP1 activity might only lead to overexpression of ZYG-1 beginning with the secondcentrioleduplicationevent.Thishoweverseemsunlikely,aswehaveshownthatlossofPP1 activityresultsinelevatedZYG-1levelseveninthefirstcellcycle(Fig4C&D),andthatitcansuppress thefailureofthefirstduplicationeventinzyg-1(it25)embryos(Fig1C&D,2B,3G).Anotherpossibility is that the critical period for exposure to elevated ZYG-1 is one cell cycle prior to the actual overduplicationevent.Thusthespermcentrioles,whichpresumablyfirstencounterelevatedZYG-1inthe zygote, would only over-duplicate during the second centriole cycle. This model is however inconsistent with previous work showing that when zyg-1 is overactive in the male germ line, centrioleswillover-duplicateduringspermatogenesis,butwhenthesecentriolesareintroducedinto awild-typeeggstheyduplicatenormally[61].Thusthepriorexposureofspermcentriolestoelevated ZYG-1activitydoesnotprogramthemtoover-duplicateinthezygote.Finally,athirdandmorelikely possibilityisthatthecentriolesneedtobeexposedtoelevatedZYG-1fortwoconsecutivecellcycles beforetheywillover-duplicate.Thussperm-derivedcentriolesarefirstexposedtoelevatedZYG-1in the zygote, but do not overduplicate until the second cell cycle, after they have been exposed to elevatedZYG-1activityfortwoconsecutivecellcycles.Thereisprecedentforthistypeofregulation; duringcentriolematurationinhumancells,Plk1activityisneededoverthecourseoftwosuccessive 19 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. cellcyclesinorderforcentriolestofullymatureandbecomecapableoforganizingPCMandserving as basal bodies [62]. Thus, this mode of regulation might be a common feature of how polo like kinasesoperate,atleastwhenitcomestothecontrolofcentriolefunction. WhydoincreasedZYG-1levelsleadtocentrioleoverduplicationintheC.elegansembryo?Normally, levelsofcentriole-associatedPlk4peakinmitosisbeforebeingreducedtoasinglefocusmarkingthe site of procentriole assembly at the G1/S transition [8,20,21]. Centrosome-associated ZYG-1 levels also peak in mitosis [63] and we speculate that elevated ZYG-1 levels prevent it from becoming restrictedtoasinglefocusduringtheensuingcellcycle,leadingtotheassemblyofmultipledaughter centrioles. Ensuring that only a single ZYG-1/Plk4 focus is maintained seems to be a key regulatory step in centriole duplication, however our understanding of how this is achieved remains limited. Clearly excess ZYG-1/Plk4 is sufficient to subvert the normal controls. Since Plk4/ZYG-1 play an important role in the recruitment of SAS-5/ana2/STIL and SAS-6 at the initiation of procentriole assembly[13,14]weenvisionelevatedZYG-1levelsmayberequiredatthistime.Howeverwhether continualexposuretoelevatedZYG-1throughtwocellcyclesisrequiredforoverduplicationremains anopenquestion. Insummary,wehaveidentifiedI-2SZY-2,SDS-22,andPP1asnovelregulatorsofcentrioleduplication. The involvement of PP1 in regulating centriole duplication has not previously been described, but interestingly centrosome amplification was reported after depletion of I-2 from human cells, suggestingthatthefunctionofPP1inregulatingcentrioleduplicationmayindeedbeconserved[31]. We show that the key function of PP1 is in limiting the availability of ZYG-1. When PP1 activity is decreased, excessive accumulation of ZYG-1 leads to the formation of extra daughter centrioles in association with a single mother, resulting in centriole amplification. Appropriate regulation of PP1 activityisthereforecrucialtomaintainingcorrectcentrosomenumbers.Althoughtherequirementfor PP1inchromosomesegregationduetoitsfunctionatthekinetochoresiswelldocumented,ourwork suggestsanovelrequirementforPP1inthemaintenanceofgenomestabilitybyregulatingcentriole duplication. 20 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. ExperimentalProcedures WormStrainsandRNAi All worm strains were maintained at 20°C on MYOB plates seeded with OP50. The strains used in these experiments are listed in supplemental table 1. To monitor the effect of the sds-22(bs9) mutation on centriole duplication sds-22(bs9) homozygotes were selected from the OC626 strain usingthevisibledpymarkerwhichiscloselylinkedtothesds-22gene.RNAiofSZY-2wascarriedout by soaking worms in dsRNA [64], all other RNAi experiments were carried out by feeding worms bacteriaexpressingdsRNAaspreviouslydescribed[65].Briefly,bacteriacontainingtheRNAiconstruct were grown overnight and seeded onto MYOB plates supplemented with 25ug/ml ampicillin and 1mMIPTG.FordoubleRNAia50:50mixofovernightculturesofthetwobacterialstrainswasplated. WormswereplacedonRNAiattheL4stagefor28hbeforeanalysis.Thesequencecontainedinthe GSP-1 RNAi construct was evaluated using the Clone mapper tool (http://bioinformatics.lif.univmrs.fr/RNAiMap/index.html)[66], which confirmed likely specificity for only the gsp-1 gene. For controlRNAiweusedansmd-1-containingvector. ImmunoprecipitationandWesternBlotting Embryonicextractsforwesternblotswerepreparedandanalyzedasdetailedpreviously[59].Whole worm extracts for the immunoprecipitation (IP) experiments were made according to [67]. Total proteinconcentrationwasdeterminedusingtheBioradProteinAssayDyeReagent(Bio-Rad).1.6mg of total protein from N2 worms was used for performing each IP. Briefly, for each IP, 30 µl of Dynabeads Protein A (Life Technologies) were incubated with 10 µg of each respective antibody at 4°Cfor2hours.Beadswerewashedthreetimesinwormlysisbuffer(50mMHEPES(pH7.4),1mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% Glycerol, 0.05% NP-40), re-suspended in 2X Laemmli Buffer (Bio-Rad), boiled at 100°C for 2 minutes and analyzed by western blotting. The following antibodies/reagents were used in this study: polyclonal SZY-2 antibodies were raised and purified againsttheentireszy-2ORF(Covance);GSP-1antibodieswereraisedandpurifiedagainstthepeptide CQYQGMNSGRPAVGGGRPGTTAGKK (YenZym Antibodies LLC); ZYG-1 (Song et. al 2008), phosphohistone H3 (Abcam); GSP-2 [68]; DM1A (Sigma). Mass spectrometry analysis was carried out by the NIDDKmassspeccorefacility. 21 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. qRT-PCR RNA was extracted from wild-type and szy-2(bs4) embryos, DNase treated and cDNA made using a superscriptIIIfirststrandsynthesiskit(Invitrogen).Forward(ACAGTACGCGGAAGAAATGG)andreverse (CACAGCAACCATCTTTTGGA)primerswereusedtoamplifyzyg-1.Primersagainstama-1wereusedas acontrol[69].qRTreactionsusediQSYBRgreensupermix(Bio-Rad)asdirectedbythemanufacturer. Imaging Fixation and staining of embryos was carried out as described previously [36]. The following antibodieswereusedata1/1000dilution:DM1A(Sigma),phospho-histoneH3(Abcam),ZYG-1[19], SPD-2 [50] and anti-SAS-4 (Song et. al 2008). For live and fixed imaging we used a spinning disk confocal microscope which has been described previously [61]. To determine whether PP1 affects translation of the ZYG-1 transcript we shifted worms carrying the reporter construct to 25°C as L4s andimagedembryosthenextday.IntensitiesofchromatinGFPatfirstmetaphaseweremeasured usingMetamorph.LevelsofZYG-1orSPD-2atthecentrosomeweredeterminedbyquantificationof average pixel intensity at the centrosome. Maximal projections of the centrosome were used for quantification of fluorescence in ImageJ 1.40g and background fluorescence was subtracted. Centrosomefluorescencewasnormalizedtocontrolssuchthatcontrolintensityis1.Forstructured illuminationmicroscopy,embryoswereimmuno-labeledasusualandmountedinVectashield(Vector Laboratories, Inc.). Samples were imaged with a DeltaVision OMX4 SIM Imaging System (Applied Precision). Acknowledgements Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Deletion strains were provided by the National BioResource Project (Japan).WethankM.ColaiacovoforC.elegansGSP-2antibody,andE.Andersonforassistancewith massspectrometry. Abbreviations PP1:proteinphosphatase1 22 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. References 1. FuJ,HaganIM,GloverDM.Thecentrosomeanditsduplicationcycle.ColdSpringHarbPerspect Biol.2015;7:a015800.doi:10.1101/cshperspect.a015800 2. HabedanckR,StierhofY-D,WilkinsonCJ,NiggEA.ThePolokinasePlk4functionsincentriole duplication.NatCellBiol.2005;7:1140–1146.doi:10.1038/ncb1320 3. StrnadP,LeidelS,VinogradovaT,EuteneuerU,KhodjakovA,GönczyP.RegulatedHsSAS-6 levelsensureformationofasingleprocentriolepercentrioleduringthecentrosome duplicationcycle.DevCell.2007;13:203–213.doi:10.1016/j.devcel.2007.07.004 4. ArquintC,SonnenKF,StierhofY-D,NiggEA.Cell-cycle-regulatedexpressionofSTILcontrols centriolenumberinhumancells.JCellSci.2012;125:1342–1352.doi:10.1242/jcs.099887 5. PeelN,StevensNR,BastoR,RaffJW.Overexpressingcentriole-replicationproteinsinvivo inducescentrioleoverduplicationanddenovoformation.CurrBiol.2007;17:834–843. doi:10.1016/j.cub.2007.04.036 6. Rodrigues-MartinsA,RiparbelliM,CallainiG,GloverDM,Bettencourt-DiasM.Revisitingthe roleofthemothercentrioleincentriolebiogenesis.Science.2007;316:1046–1050. doi:10.1126/science.1142950 7. DzhindzhevNS,TzolovskyG,LipinszkiZ,SchneiderS,LattaoR,FuJ,etal.Plk4phosphorylates Ana2totriggerSas6recruitmentandprocentrioleformation.CurrBiol.2014;24:2526–2532. doi:10.1016/j.cub.2014.08.061 8. OhtaM,AshikawaT,NozakiY,Kozuka-HataH,GotoH,InagakiM,etal.Directinteractionof Plk4withSTILensuresformationofasingleprocentrioleperparentalcentriole.NatCommun. 2014;5:5267.doi:10.1038/ncomms6267 9. KratzA-S,BärenzF,RichterKT,HoffmannI.Plk4-dependentphosphorylationofSTILisrequired forcentrioleduplication.BiolOpen.2015;4:370–377.doi:10.1242/bio.201411023 10. MoyerTC,ClutarioKM,LambrusBG,DaggubatiV,HollandAJ.BindingofSTILtoPlk4activates kinaseactivitytopromotecentrioleassembly.JCellBiol.2015;209:863–878. doi:10.1083/jcb.201502088 11. KitagawaD,FlückigerI,PolanowskaJ,KellerD,ReboulJ,GönczyP.PP2APhosphataseActs uponSAS-5toEnsureCentrioleFormationinC. elegansEmbryos.DevCell.2011;20: 550–562.doi:10.1016/j.devcel.2011.02.005 12. vanBreugelM,HironoM,AndreevaA,YanagisawaH-A,YamaguchiS,NakazawaY,etal. StructuresofSAS-6SuggestItsOrganizationinCentrioles.Science.2011. doi:10.1126/science.1199325 13. DelattreM,CanardC,GönczyP.SequentialproteinrecruitmentinC.eleganscentriole 23 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. formation.CurrBiol.2006;16:1844–1849.doi:10.1016/j.cub.2006.07.059 14. LettmanMM,WongYL,ViscardiV,NiessenS,ChenS-H,ShiauAK,etal.DirectbindingofSAS-6 toZYG-1recruitsSAS-6tothemothercentrioleforcartwheelassembly.DevCell.2013;25:284– 298.doi:10.1016/j.devcel.2013.03.011 15. SillibourneJE,TackF,VloemansN,BoeckxA,ThambirajahS,BonnetP,etal. Autophosphorylationofpolo-likekinase4anditsroleincentrioleduplication.MolBiolCell. 2010;21:547–561.doi:10.1091/mbc.E09-06-0505 16. HollandAJ,LanW,NiessenS,HooverH,ClevelandDW.Polo-likekinase4kinaseactivitylimits centrosomeoverduplicationbyautoregulatingitsownstability.JCellBiol.2010;188:191–198. doi:10.1083/jcb.200911102 17. GuderianG,WestendorfJ,UldschmidA,NiggEA.Plk4trans-autophosphorylationregulates centriolenumberbycontrollingbetaTrCP-mediateddegradation.JCellSci.2010;123:2163– 2169.doi:10.1242/jcs.068502 18. RogersGC,RusanNM,RobertsDM,PeiferM,RogersSL.TheSCFSlimbubiquitinligase regulatesPlk4/Saklevelstoblockcentriolereduplication.JCellBiol.2009;184:225–239. doi:10.1083/jcb.200808049 19. PeelN,DoughertyM,GoeresJ,LiuY,O'ConnellKF.TheC.elegansF-boxproteinsLIN-23and SEL-10antagonizecentrosomeduplicationbyregulatingZYG-1levels.JCellSci.2012;125: 3535–3544.doi:10.1242/jcs.097105 20. KimT-S,ParkJ-E,ShuklaA,ChoiS,MuruganRN,LeeJH,etal.HierarchicalrecruitmentofPlk4 andregulationofcentriolebiogenesisbytwocentrosomalscaffolds,Cep192andCep152. ProceedingsoftheNationalAcademyofSciences.2013;110:E4849–57. doi:10.1073/pnas.1319656110 21. SonnenKF,GabryjonczykA-M,AnselmE,StierhofY-D,NiggEA.HumanCep192andCep152 cooperateinPlk4recruitmentandcentrioleduplication.JCellSci.2013;126:3223–3233. doi:10.1242/jcs.129502 22. ArquintC,GabryjonczykA-M,ImsengS,BöhmR,SauerE,HillerS,etal.STILbindingtoPolo-box 3ofPLK4regulatescentrioleduplication.Elife.2015;4.doi:10.7554/eLife.07888 23. KlebbaJE,BusterDW,McLamarrahTA,RusanNM,RogersGC.Autoinhibitionandrelief mechanismforPolo-likekinase4.ProceedingsoftheNationalAcademyofSciences.2015;112: E657–66.doi:10.1073/pnas.1417967112 24. KirchnerJ,GrossS,BennettD,AlpheyL.Essential,overlappingandredundantrolesofthe Drosophilaproteinphosphatase1alphaand1betagenes.Genetics.2007;176:273–281. doi:10.1534/genetics.106.069914 25. SassaT,Ueda-OhbaH,KitamuraK-I,HaradaS-I,HosonoR.RoleofCaenorhabditiselegans proteinphosphatasetype1,CeGLC-7beta,inmetaphasetoanaphasetransitionduring 24 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. embryonicdevelopment.ExpCellRes.2003;287:350–360. 26. WuJ-C,GoAC,SamsonM,CintraT,MirsoianS,WuTF,etal.Spermdevelopmentandmotility areregulatedbyPP1phosphatasesinCaenorhabditiselegans.Genetics.2012;190:143–157. doi:10.1534/genetics.111.135376 27. VirshupDM,ShenolikarS.Frompromiscuitytoprecision:proteinphosphatasesgeta makeover.MolCell.2009;33:537–545.doi:10.1016/j.molcel.2009.02.015 28. HuangFL,GlinsmannWH.Separationandcharacterizationoftwophosphorylasephosphatase inhibitorsfromrabbitskeletalmuscle.EurJBiochem.1976;70:419–426. 29. TungHY,WangW,ChanCS.RegulationofchromosomesegregationbyGlc8p,astructural homologofmammalianinhibitor2thatfunctionsasbothanactivatorandaninhibitorofyeast proteinphosphatase1.MolCellBiol.1995;15:6064–6074. 30. LiM,SatinoverDL,BrautiganDL.Phosphorylationandfunctionsofinhibitor-2familyof proteins.Biochemistry.2007;46:2380–2389.doi:10.1021/bi602369m 31. WangW,StukenbergPT,BrautiganDL.Phosphataseinhibitor-2balancesproteinphosphatase1 andauroraBkinaseforchromosomesegregationandcytokinesisinhumanretinalepithelial cells.MolBiolCell.2008;19:4852–4862.doi:10.1091/mbc.E08-05-0460 32. WangW,CronmillerC,BrautiganDL.Maternalphosphataseinhibitor-2isrequiredforproper chromosomesegregationandmitoticsynchronyduringDrosophilaembryogenesis.Genetics. 2008;179:1823–1833.doi:10.1534/genetics.108.091959 33. PoschM,KhoudoliG,SwiftS,KingE,DeLucaJ,SwedlowJ.Sds22regulatesauroraBactivityand microtubule–kinetochoreinteractionsatmitosis.JCellBiol.2010;191:61. 34. MeraldiP,NiggEA.Centrosomecohesionisregulatedbyabalanceofkinaseandphosphatase activities.JCellSci.2001;114:3749–3757. 35. HelpsNR,LuoX,BarkerHM,CohenPT.NIMA-relatedkinase2(Nek2),acell-cycle-regulated proteinkinaselocalizedtocentrosomes,iscomplexedtoproteinphosphatase1.BiochemJ. 2000;349:509–518. 36. KempCA,SongMH,AddepalliMK,HunterG,O'ConnellK.Suppressorsofzyg-1define regulatorsofcentrosomeduplicationandnuclearassociationinCaenorhabditiselegans. Genetics.2007;176:95–113.doi:10.1534/genetics.107.071803 37. HsuJY,SunZW,LiX,ReubenM,TatchellK,BishopDK,etal.Mitoticphosphorylationofhistone H3isgovernedbyIpl1/aurorakinaseandGlc7/PP1phosphataseinbuddingyeastand nematodes.Cell.2000;102:279–291. 38. QianJ,LesageB,BeullensM,VanEyndeA,BollenM.PP1/Repo-mandephosphorylatesmitotic histoneH3atT3andregulateschromosomalauroraBtargeting.CurrBiol.2011;21:766–773. doi:10.1016/j.cub.2011.03.047 25 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. 39. KaitnaS,PasierbekP,JantschM,LoidlJ,GlotzerM.TheauroraBkinaseAIR-2regulates kinetochoresduringmitosisandisrequiredforseparationofhomologousChromosomesduring meiosis.CurrBiol.2002;12:798–812. 40. OhkuraH,YanagidaM.S.pombegenesds22+essentialforamidmitotictransitionencodesa leucine-richrepeatproteinthatpositivelymodulatesproteinphosphatase-1.Cell.1991;64: 149–157. 41. StoneEM,YamanoH,KinoshitaN,YanagidaM.Mitoticregulationofproteinphosphatasesby thefissionyeastsds22protein.CurrBiol.1993;3:13–26. 42. MacKelvieSH,AndrewsPD,StarkMJ.TheSaccharomycescerevisiaegeneSDS22encodesa potentialregulatorofthemitoticfunctionofyeasttype1proteinphosphatase.MolCellBiol. 1995;15:3777–3785. 43. RogersE,BishopJD,WaddleJA,SchumacherJM,LinR.TheaurorakinaseAIR-2functionsinthe releaseofchromosomecohesioninCaenorhabditiselegansmeiosis.JCellBiol.2002;157:219– 229.doi:10.1083/jcb.200110045 44. SchumacherJM,GoldenA,DonovanPJ.AIR-2:AnAurora/Ipl1-relatedproteinkinaseassociated withchromosomesandmidbodymicrotubulesisrequiredforpolarbodyextrusionand cytokinesisinCaenorhabditiselegansembryos.JCellBiol.1998;143:1635–1646. 45. SatinoverDL,LeachCA,StukenbergPT,BrautiganDL.ActivationofAurora-Akinasebyprotein phosphataseinhibitor-2,abifunctionalsignalingprotein.ProcNatlAcadSciUSA.2004;101: 8625–8630.doi:10.1073/pnas.0402966101 46. HannakE,KirkhamM,HymanAA,OegemaK.Aurora-Akinaseisrequiredforcentrosome maturationinCaenorhabditiselegans.JCellBiol.2001;155:1109–1116. doi:10.1083/jcb.200108051 47. GloverDM,LeibowitzMH,McLeanDA,ParryH.Mutationsinaurorapreventcentrosome separationleadingtotheformationofmonopolarspindles.Cell.1995;81:95–105. 48. YamashiroS,YamakitaY,TotsukawaG,GotoH,KaibuchiK,ItoM,etal.Myosinphosphatasetargetingsubunit1regulatesmitosisbyantagonizingpolo-likekinase1.DevCell.2008;14:787– 797.doi:10.1016/j.devcel.2008.02.013 49. TsouM-FB,WangW-J,GeorgeKA,UryuK,StearnsT,JallepalliPV.Polokinaseandseparase regulatethemitoticlicensingofcentrioleduplicationinhumancells.DevCell.2009;17:344– 354.doi:10.1016/j.devcel.2009.07.015 50. KempCA,KopishKR,ZipperlenP,AhringerJ,O'ConnellKF.Centrosomematurationand duplicationinC.elegansrequirethecoiled-coilproteinSPD-2.DevCell.2004;6:511–523. 51. PelletierL,OzlüN,HannakE,CowanC,HabermannB,RuerM,etal.TheCaenorhabditis eleganscentrosomalproteinSPD-2isrequiredforbothpericentriolarmaterialrecruitmentand centrioleduplication.CurrBiol.2004;14:863–873.doi:10.1016/j.cub.2004.04.012 26 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. 52. DeckerM,JaenschS,PozniakovskyA,ZinkeA,O'ConnellKF,ZachariaeW,etal.Limiting amountsofcentrosomematerialsetcentrosomesizeinC.elegansembryos.CurrBiol.2011;21: 1259–1267.doi:10.1016/j.cub.2011.06.002 53. SongMH,AravindL,Müller-ReichertT,O'ConnellKF.TheconservedproteinSZY-20opposes thePlk4-relatedkinaseZYG-1tolimitcentrosomesize.DevCell.2008;15:901–912. doi:10.1016/j.devcel.2008.09.018 54. MillerJG,LiuY,WilliamsCW,SmithHE,O'ConnellKF.TheE2F-DP1TranscriptionFactor ComplexRegulatesCentrioleDuplicationinCaenorhabditiselegans.G3(Bethesda).2016;6: 709–720.doi:10.1534/g3.115.025577 55. BastoR,BrunkK,VinadogrovaT,PeelN,FranzA,KhodjakovA,etal.Centrosomeamplification caninitiatetumorigenesisinflies.Cell.2008;133:1032–1042.doi:10.1016/j.cell.2008.05.039 56. FuJ,GloverDM.Structuredilluminationoftheinterfacebetweencentrioleandperi-centriolar material.OpenBiol.2012;2:120104.doi:10.1098/rsob.120104 57. SonnenKF,SchermellehL,LeonhardtH,NiggEA.3D-structuredilluminationmicroscopy providesnovelinsightintoarchitectureofhumancentrosomes.BiolOpen.2012;1:965–976. doi:10.1242/bio.20122337 58. BrownleeCW,KlebbaJE,BusterDW,RogersGC.TheProteinPhosphatase2Aregulatory subunitTwinsstabilizesPlk4toinducecentrioleamplification.JCellBiol.2011;195:231–243. doi:10.1083/jcb.201107086 59. SongMH,LiuY,AndersonDE,JahngWJ,O'ConnellKF.ProteinPhosphatase2A-SUR-6/B55 RegulatesCentrioleDuplicationinC.elegansbyControllingtheLevelsofCentrioleAssembly Factors.DevCell.2011;20:563–571.doi:10.1016/j.devcel.2011.03.007 60. EgloffMP,JohnsonDF,MoorheadG,CohenPT,CohenP,BarfordD.Structuralbasisforthe recognitionofregulatorysubunitsbythecatalyticsubunitofproteinphosphatase1.EMBOJ. EMBOPress;1997;16:1876–1887.doi:10.1093/emboj/16.8.1876 61. PetersN,PerezDE,SongMH,LiuY,Müller-ReichertT,CaronC,etal.Controlofmitoticand meioticcentrioleduplicationbythePlk4-relatedkinaseZYG-1.JCellSci.2010;123:795–805. doi:10.1242/jcs.050682 62. KongD,FarmerV,ShuklaA,JamesJ,GruskinR,KiriyamaS,etal.Centriolematurationrequires regulatedPlk1activityduringtwoconsecutivecellcycles.JCellBiol.RockefellerUniversity Press;2014;206:855–865.doi:10.1083/jcb.201407087 63. O'ConnellKF,CaronC,KopishKR,HurdDD,KemphuesKJ,LiY,etal.TheC.eleganszyg-1gene encodesaregulatorofcentrosomeduplicationwithdistinctmaternalandpaternalrolesinthe embryo.Cell.2001;105:547–558. 64. FernandezAG,GunsalusKC,HuangJ,ChuangL-S,YingN,LiangH-L,etal.Newgeneswithroles intheC.elegansembryorevealedusingRNAiofovary-enrichedORFeomeclones.GenomeRes. 27 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. 2005;15:250–259.doi:10.1101/gr.3194805 65. KamathRS,FraserAG,DongY,PoulinG,DurbinR,GottaM,etal.Systematicfunctionalanalysis oftheCaenorhabditiselegansgenomeusingRNAi.Nature.2003;421:231–237. doi:10.1038/nature01278 66. ThakurN,PujolN,TichitL,EwbankJJ.Clonemapper:anonlinesuiteoftoolsforRNAi experimentsinCaenorhabditiselegans.G3(Bethesda).GeneticsSocietyofAmerica;2014;4: 2137–2145.doi:10.1534/g3.114.013052 67. ZaninE,DumontJ,GassmannR,CheesemanI,MaddoxP,BahmanyarS,etal.Affinity purificationofproteincomplexesinC.elegans.MethodsCellBiol.2011;106:289–322. doi:10.1016/B978-0-12-544172-8.00011-6 68. TzurYB,EgydiodeCarvalhoC,NadarajanS,VanBostelenI,GuY,ChuDS,etal.LAB-1targets PP1andrestrictsAuroraBkinaseuponentranceintomeiosistopromotesisterchromatid cohesion.PLoSBiol.2012;10:e1001378.doi:10.1371/journal.pbio.1001378 69. WangH,SpangA,SullivanMA,HryhorenkoJ,HagenFK.Theterminalphaseofcytokinesisin theCaenorhabditiselegansearlyembryorequiresproteinglycosylation.IntRevCytol.2005;16: 4202–4213.doi:10.1091/mbc.E05-05-0472 SupportingInformation SupplementalFigureS1.Centriolebehaviorinvariousgeneticbackgrounds.Schematicshowinghow centriolesbehaveandhowtheirnumbersareestablishedateachcellstagethroughthefirstthreecell cyclesinA)wild-typeembryos,B)zyg-1(it25)mutantembryosandinC)embryoswithreducedPP1 function. Supplemental Figure S2. RNAi of PP1 regulators in zyg-1(it25) worms. Quantificationofembryonic viability among the progeny of worms grown at the semi-permissive temperature of 24°C and depleted for the indicated PP1 regulators. smd-1(RNAi) targets a nonessential gene and serves as a negativecontrol.Onlyreductionofsds-22ledtosubstantialrescueofzyg-1(it25)lethality. SupplementalFigureS3.LocalizationofI-2SZY-2,PP1βGSP-1andSDS-22.GFPfusionsoftheindicated proteinwereexpressedintheembryo.I-2SZY-2Isenrichedinthenuclei.Pronucleimeetingduringthe firstcellcycleisshown.PP1βGSP-1isenrichedonthechromatinthroughoutmitosis.Anaphaseofthe 28 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. firstcellcycleisshown.SDS-22isweaklylocalizedtothespindle.Metaphaseofthefirstcellcycleis shown. SupplementalFigureS4.SummaryofImmunoprecipitation-MassSpectrometryResults.Eachofthe indicatedproteinswasimmunoprecipitatedfrom wormextractsandco-purifyingproteinsidentified bymassspec.Shownarethetopfivehitsbasedonpeptidenumber.IncaseswherePP1GSP-1,SDS-22 or I-2SZY-2were not among the top hits, they are also shown along with their rank and number of identifyingpeptides. Supplemental Figure S5. Loss of SDS-22 does not elevate phospho-histone H3 levels in mitotic chromatin. A) Western blot showing elevated total phospho-histone levels present in extract from mixedstagesds-22(b9/tm5187)embryos.Thiselevationislikelyduetoanincreaseinthelengthof mitosis in sds-22 mutant embryos as B) quantitative immunofluorescence microscopy shows that mitoticchromatininsds-22(bs9/tm5187)embryosisnotenrichedforphospho-histoneH3relativeto thewildtype,andinfact,appearsreduced.(a.u.=arbitraryunits) AIR-2 SupplementalFigureS6.AuroraAAIR-1,AuroraB ,andPlk-1donotopposePP1inthecontrolof centriole duplication. Each of the specified kinases were RNAi depleted in zyg-1(i25); szy-2(bs4) wormsandcentrioleduplicationmonitoredbymicroscopy.A)Quantificationofcentrioleduplication. Numbers above bars indicate the percentage of successful centriole duplication events and the numberwithinthebarsindicatethenumberofeventsscored.B-D)Representativestillsfromtimelapserecordingsofzyg-1(it25);szy-2(bs4)embryosexpressingGFP::tubulinandmCherry::histoneand treated with control RNAi or RNAi against one of the three indicated mitotic kinases. B) Embryos treatedwithcontrolRNAiduplicatecentrioles,proceedtothe2cellstage,andbuildbipolarspindles. C) Embryos treated with air-2 RNAi duplicate centrioles, but fail in chromosome segregation, thus display4centrosomesassociatedwithasingleenlargednucleus.D)Embryostreatedwithplk-1RNAi showavarietyofcelldivisiondefectsyetpresentwith4centrosomesindicatingcentrioleduplication hasoccurred.E&F)Representativestillsfromtime-lapserecordingsofzyg-1(it25);szy-2(bs4)embryos expressingGFP::SPD-2andtreatedwithcontrolRNAi(E)orRNAiagainstair-1(F).CentriolesfromF areenlargedina&b.Embryosareattheearly2-cellstageandduplicatedcentriolesarevisible(arrow heads).Note,inthisexampleonlyonecentrioleofthecontrolembryohasduplicated. 29 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Supplemental Figure S7. Centrosome-associated SPD-2 levels are elevated in szy-2(bs4) mutants, butthisdoesnotappeartocontributetorescueofthezyg-1(it25)phenotype.A)AverageGFP::SPD2 levels at the centrosome during the first cell cycle were calculated and normalized to control. B) Average levels of endogenous SPD-2 at the centrosome during the first metaphase in szy-2(bs4) embryoswerenormalizedtocontrols.C&D)Representativewildtypeandszy-2(bs4)embryosstained forDNA(blue),tubulin(red)andSPD-2(green).SPD-2stainingatcentrosomesisenlargedinaandb. E&F)ComparisonofSPD-2::GFPlevelsatthecentrosomeinanembryoexpressingacodon-optimized version of SPD-2::GFP (OE=overexpression) (E) and a strain expressing GFP::SPD-2 from the native sequence (F). G) Quantification of embryonic viability among the indicated strains to determine whetheroverexpressionofSPD-2issufficienttorescuethezyg-1(it25)phenotype.Ineachcasen=15 worms, >1000 embryos. H) GFP::SPD-2 levels at the centrosome in zyg-1(it25); szy-2(bs4) embryos treatedwithnegativecontrolsmd-1(RNAi)orair-1(RNAi). SupplementalFigureS8.SAS-6levelsarenotalteredbyreductionofPP1activity.A)Westernblot demonstratingequivalenttotallevelsofSAS-6inwild-typeandszy-2(bs4)embryos.B)Quantitation ofwesternblotdata(n=2).C)Averagelevelsofcentrosome-localizedGFP::SAS-6inwild-typeandszy2(bs4)embryosweredeterminedthroughoutthefirstcellcycle.Valuesshownarenormalizedtothe averagewild-typevalueateachstage.Errorbarsrepresentstandarderror. SupplementalFigureS9.Centriolescontinuetoover-duplicateinolderembryos.Selectframesfrom a time-lapse recording of an sds-22(bs9/tm5187) embryo expressing GFP::SPD-2 and mCherry::histone. The arrowhead shows a spindle pole (frame 18:00) giving rise to multiple centrosomesduringthenextcellcycle(fram28:00).Whitearrowsindicatecentrosomesthatbecome inactiveduringtheroundofdivisionfollowingtheirbirth(frames10:00and18:00)butbecomeactive againduringthenextcellcycle(frames22:00-34:00).Notethatbothcentrosomeswerenotalways visibleintheseframes. SupplementaryMovies MovieS1-Normalcentrioleduplicationinawild-typeembryoexpressingGFP::tubulinand mCherry::histone.Notebipolarspindleformationatthetwo-cellstage. 30 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. MovieS2-Centrioleduplicationinazyg-1(it25)mutantembryoexpressingGFP::SPD-2and mCherry::histone.Notemonopolarspindleformationatthetwo-cellstage. MovieS3-Centrioleduplicationinzyg-1(it25);szy-2(bs4)doublemutantembryoexpressing GFP::tubulinandmCherry::histone.Noterestorationofbipolarspindleformationatthetwo-cellstage. MovieS4-AnaphasebridgescanbeseeninembryosexpressingGFP::histoneandmCherry::SPD-2 whengsp-1andgsp-2areco-depletedbyRNAi. MovieS5-EmbryosexpressingGFP::histoneandmCherry::SPD-2produceextracentrosomesatthe4cellstagewhengsp-1andgsp-2areco-depletedbyRNAi. MovieS6-Extracentrosomesarepresentatthefour-cellstageinsds-22(bs9/tm5187)embryos expressingGFP::SPD-2andmCherry::histone. MovieS7-Insds-22(bs9/tm5187)embryosexpressingGFP::SPD-2andmCherry::histone,extra centrosomescontinuetobeproducedafterthe4-cellstage.TheextracentrosomesrecruitPCMand participateinspindleassembly. MovieS8-Theextracentrosomesdetectedingsp-1(RNAi);gsp-2(RNAi)embryosexpressing GFP::histoneandmCherry::SPD-2functionasmicrotubule-organizingcentersandgiveriseto multipolarspindles. SupplementalTable1C.elegansstrainsusedinthisstudy Strainname OC467 OC192 OC227 OC14 OC192 OC199 OC367 OC429 OC448 OC469 OC573 Genotype szy-2(tm3972)/hT2[bli-4(e937)let?(q782)qIs48](I;III) szy-2(bs4)III zyg-1(it25)II;szy-2(bs4)III zyg-1(it25)II szy-2(bs4)III zyg-1(it25)sds-22(bs9)II zyg-1(it25)II;szy-2(bs4)unc-119(ed3)III;ltIs37[pAA64:unc-119(+)pie1p::mcherry::his58];ltIs25[pAZ132:unc-119(+)pie-1p::gfp::tba-2] dpy-17(e164)szy-2(bs4)III;bsIs2[pCK5.5::unc-119(+)pie-1p::gfp::spd-2] zyg-1(it25)II;ltIs37[pAA64:unc-119(+)pie-1p::mcherry::his-58]; ltIs25[pAZ132:unc-119(+)pie-1p::gfp::tba-2] zyg-1(it25)II;szy-2(tm3972)III;ltIs37[pAA64:unc-119(+)pie1p::mcherry::his-58];ltIs25[pAZ132:unc-119(+)pie-1p::gfp::tba-2] sds-22(tm5187)/mIn1[dpy-10(e128)mIs14(myo-2::gfp)]II 31 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. OC156 OC381 OC626 OC591 OC92 NIN33 OC770 OC779 OC723 OC835 OC547 OC484 OC426 zyg-1(it25)II;bsIs2[pCK5.5::unc-119(+)pie-1p::gfp::spd-2];unc119(ed3)ruIs32[unc-119(+)pie-1p::gfp::his-58]III zyg-1(it25)II;gsp-2(tm301)/hT2[bli-4(e937)let?(q782)qIs48](I;III); ltIs37[pAA64:unc-119(+)pie-1p::mcherry::his-58];ltIs25[pAZ132:unc119(+)pie-1p::gfp::tba-2] sds-22(bs9)/sds-22(tm5187)II;bsIs2[pCK5.5:unc-119(+)pie-1p::gfp::spd2];ltIs37[pAA64:unc-119(+)pie-1p::mcherry::his-58] zyg-1(it25);[unc-119(+)pie-1-GFP::SPD-2OE]InsertionfromDeckeretal, 2011 unc-119(ed4)III;bsIs2[pCK5.5:unc-119(+)pie-1p::gfp::spd-2] szy-2(bs4)unc119(ed3)III;fem-1(hc17)IV;ItIs33[pOD224:unc-119(+)pie1p::gfp::tev::stag::sas-6;] bsSi15[pKO109:spd-2p::spd-2::mCherry::spd-23'-utr,unc-119(+)]I;szy2(tm3972)III bsSi15[pKO109:spd-2p::spd-2::mCherry::spd-23'-utr,unc-119(+)]I; bsSi30[pCW9:ZC504.3p::sfgfp::his-58::ZC504.33’-utr,unc-119(+)]II bsSi9[pKO113:unc-119(+)zyg-1p::gfp::his-58::3'utrzyg-1]I;unc119(ed3)III bsIs9[pKO113:unc-119(+)zyg-1p-gfp::his-58-zyg-13'utr]I;szy-2(tm3972) III unc-119(ed3)III: bsIs33 [pNP121: pie-1promoter-gfp-sds-22-pie-1UTR unc-119(+)]/+ unc-119(ed3) III; BsIs18[pNP97: pie1promoter-gfp-gsp-1genomicpie1UTR unc-119(+)] unc119(ed3)III; pNP71: bsIs52[pie-1promoter-gfp::szy-2-pie-1UTR unc119(+)] 32 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 1 A W T tm3972 deletion sz y2 szy-2 (b s4 ) B 100bp 38 SZY-2 szy-2(bs4) G-A 28 α-tubulin C D 100 70 60 50 40 30 20 39 (tm -2 zy ;s 5) it2 1( g- zy P-H3 ) 32 72 ) Ai (R -2 zy it2 1( g- 32 N N ;s 5) ;c 5) it2 1( gzy 20 R l( tro on ;s 5) it2 bs y2( merge 20 Ai ) s4 2( y1( gzy α-tubulin szy-2(bs4) P-H3 sz w ild ty pe E 4) zyg-1(it25); szy-2(bs4) Wild type F 20 (b bs 5) sz it2 1( gzy zyg-1(it25) 74 ) 22 zy >50 0 -2 10 zy wild type 80 4) % centriole duplication 90 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 2 A sds-22 tm5187 deletion 100bp sds-22(bs9) G224S B 100 % Centriole duplication 90 80 70 60 50 40 30 20 10 0 20 28 20 zyg-1(it25) sds-22(bs9) 20 zyg-1(it25); sds-22 - (bs9) Genotype zyg-1(it25); sds-22(RNAi) C Genotype dpy-10(e128) sds-22(bs9) dpy-10(e128) sds-22(bs9)/ + + sds-22(tm5187) sds-22(tm5187)/ + sds-22(tm5187)/szy-6(bs9) Viability # embryos 96% 245 99.2% 1016 Larval lethal n/a 77.5% 151 0% 579 bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 3 B A (A.U.) ) Ai ) Ai R 1( p- nt gs 350 PP1βGSP-1 40kDa 300 Percent knockdown 250 200 150 100 N N l( R ro 400 co Phospho-HH3 Ser10 fluorescence intensity 450 control (RNAi) 91% 40kDa PP1αGSP-2 50kDa Tubulin gsp-1/2 (RNAi) D C zyg-1(it25) zyg-1(it25); gsp-1(RNAi) E F zyg-1(it25) % centriole duplication G 100 90 80 70 60 50 40 30 20 10 0 20 20 zyg-1(it25) zyg-1(it25) gsp-2(tm301) 26 zyg-1(it25); gsp-2(tm301) Genotype IP Blot: PP1β GSP-1 Blot: I-2 SZY-2 Blot: I-2 SZY-2 Blot: SDS-22 Blot: SDS-22 S22 SD SZ I-2 lI ro nt Y2 gG ) % in pu PP Blot: PP1α GSP-2 co t( 13 1 SP - I 1β G Y2 SZ gG I-2 lI ro nt co in pu t( 13 % ) H IP zyg-1(it25); gsp-1(RNAi) bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 4 6 Normalized ZYG-1 protein level szy-2(tm3972) wild type N R 2( gs p- 1/ R gs ) ) p- 1/ 2( 2( R R N Ai Ai ) N Ai ) Normalized zyg-1 transcript level H p- 2( R p- 1( G WT gs WT Ai ) N Ai N s9 22 s- sd Ai ) /tm ) 51 87 ) F (b 4) bs 2( ysz WT 72 sd Cell Cycle Stage E p- Anaphase 73 gs Metaphase N 58 52 (b Prophase 50 54 22 94 63 59 48 63 sz 0 74 0 y- 0.4 n.s. 0.5 R 0.6 n.s. 1 gs 0.8 1.5 87 0.93 1( 1 4) 1 ** ** ** s9 1 2 bs n.s. szy-2 (bs4) 2.5 T 1.26 WT D 2( ** szy-2(bs4) W WT 1.2 0.2 1 s- Normalized ZYG -1 Fluorescence 1.51 1.4 1 2 0 ** 1.6 Normalized ZYG -1 fluorescence in prophase C 3 p- Tubulin 4 gs ZYG-1 5 51 (b s4 ) sz y2 zy g- w il d -t y pe Ai ) RN 1( co nt ro lR NA i B /tm A 1.2 1 0.8 0.6 0.4 0.2 0 WT szy-2(bs4) Genotype bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 5 A % of duplications producing supernumerary centrioles B sds-22(bs9/tm5187) sds-22(bs9)/sds-22(tm5187) szy-2(tm3972) szy-2(tm3972) 45 40 35 30 25 20 15 10 5 0 centriole cycle 1 centriole cycle 2 Timing of centrosome duplication event Ai ) D RN p1/ 2( gs co n tro l(R NA i) C PP1βGSP-1 PP1αGSP-2 Tubulin E F WT ) (R 1/ 2 G Percentage of centrosomes Ai N ) Ai gs p- 2( R N ) gs p- gs p- 1( R N Ai % of embryos showing overduplication 100 90 80 70 60 50 40 30 20 10 0 70 sds-22(bs9/tm5187) 63 60 49 50 40 37 33 30 18 20 10 0 0 1 centriole 2 centrioles >2 centrioles bioRxiv preprint first posted online Dec. 12, 2016; doi: http://dx.doi.org/10.1101/093492. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. All rights reserved. No reuse allowed without permission. Figure 6 SCF ? SDS-22 PP1 I-2SZY-2 ZYG-1 SAS-6 SAS-5 SAS-4 Centriole duplication