* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Hafiz Noordin Term Paper - Engineering Computing Facility
Cortical cooling wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Premovement neuronal activity wikipedia , lookup
Convolutional neural network wikipedia , lookup
Neuroeconomics wikipedia , lookup
Incomplete Nature wikipedia , lookup
Holonomic brain theory wikipedia , lookup
Agent-based model wikipedia , lookup
Synaptic gating wikipedia , lookup
Visual servoing wikipedia , lookup
Neural modeling fields wikipedia , lookup
Neural correlates of consciousness wikipedia , lookup
Neuroesthetics wikipedia , lookup
Metastability in the brain wikipedia , lookup
C1 and P1 (neuroscience) wikipedia , lookup
Cerebral cortex wikipedia , lookup
Biological neuron model wikipedia , lookup
Inferior temporal gyrus wikipedia , lookup
Efficient coding hypothesis wikipedia , lookup
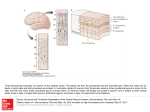
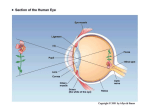
1 A Systems Biology View of Modeling the Visual Cortex Hafiz Z. Noordin Abstract— System-level understanding of biological systems has become a topic of great importance in modern science. The systems biology paradigm provides a fundamental framework for methodically modeling and simulating biological systems. Recent models can be shown to follow this structural approach, however issues regarding assumptions and functional accuracy must be considered during the process. This paper will present the systems biology perspective of modeling the visual cortex, as well as introduce and demonstrate a recently developed visual system model, Topographica. This model will be evaluated by considering the significant issues inherent to the modeling process. Index Terms — computational modeling, cortical maps, systems biology, visual cortex I. INTRODUCTION T HE emergence of systems biology as a unified approach to understanding biological systems has compelled scientists and engineers to reevaluate modern methodologies. In particular, the techniques used to characterize biological systems as mathematical models must follow a particular “Framework for Systems Biology” [1] in order to correctly adhere to the four phases of system-level understanding [2]. This definition of systems biology research implies that the modeling process should be accordingly structured in order to demonstrate the validity of the resultant model. Due to the recent availability of complete genome sequences, as well as the emergence of high-throughput measurement systems at the genetic level [2], much of the recent systems biology research has been targeted towards modeling at this low-level stage of the biological hierarchy. This paper, however, will demonstrate how the systems biology approach can be applied at a higher level, by applying the aforementioned “Framework of Systems Biology” to modeling the visual cortex. In particular, it will be shown that the concepts emphasized by the systems biology paradigm are certainly significant when considering the issues and difficulties encountered when attempting to model complex networks such as the visual cortex. The paper will begin with a brief summary of the systems biology approach, as well as an overview of the visual system Hafiz Z. Noordin is with the Institute of Biomaterials and Biomedical Engineering, University of Toronto, ON, Canada (e-mail: hafiz.noordin@ utoronto.ca). in order to provide the necessary background. A recent approach to modeling the visual cortex will then be demonstrated, and finally design issues and model evaluation from the perspective of systems biology will be discussed. II. THE SYSTEMS BIOLOGY VIEW Systems biology is made possible by recent parallel advances in high throughput experimental technologies along with advances in fields such as computer science and engineering. Because of the mass of information produced by the tools employed, systems biology is also an informational science. It aims to gather information from different hierarchical levels of the biological system and to integrate them to build predictive mathematical models of the system. This will primarily be used to offer a comprehensive and consistent body of knowledge in biology [2]. A systems level understanding is used to combine component level and dynamic information in order to determine the state of a system and how it changes. Four fundamental experimental approaches are used to do this: 1. 2. 3. 4. Structure and function determination Dynamics of the system and interplay of different components Methods to control and perturb the system Methods to design and modify the system, and forward engineering Progress has taken place in this order; however insufficient progress has been made in simulating dynamics of the systems [2]. In modeling the visual cortex, this is certainly an issue, as the difficulties lie more in the interactions of network components. The basic “Framework of Systems Biology” [1] consists of: 1. 2. 3. 4. Formulating an initial model by defining the components of the system Systematically perturbing and observing the components, either through internal or environmental means Refining the model in an iterative manner by continuously comparing predicted results and experimental observations Redesigning model structure and dynamics, as well as experimentation and validation techniques based on the 2 success of the model. These steps provide a generic approach to modeling, and hence it will be shown how this approach is applied to a recent model of the visual cortex. III. PHYSIOLOGY OF THE VISUAL SYSTEM In order to understand models of the visual cortex, it is important that the reader has some background on the physiology of the visual system, since the terminology utilized in this field tends to reflect the complex nature of the visual cortex itself. It is assumed, however, that the reader has basic knowledge of the neuron, at least at a functional level. The structure of the eye is shown in Figure 1. The eye accepts photons of light via the retina, which contains a layer of photoreceptors (rods and cones) interfacing indirectly to a layer of ganglion cells. The purpose of this interface is to transduce the light into a neural response [3]. The generated neural signal is then transmitted to the brain via the optic nerve. A key term that is used in describing this process is the concept of a receptive field. A single ganglion cell’s receptive field consists of a region of retina upon which incident light alters the firing rate of the cell. Furthermore, the receptive field contains either or both of an on-response (firing immediately after the onset of a stimulus) and off-response (firing immediately after termination of stimulus) [3]. those of the retinal ganglion cells [3]. Finally, the outputs of the LGN synapse with the primary visual cortex by means of optic radiation. An important feature of this stage of the visual pathway is that an estimated 80-90% of the inputs to the LGN are from higher cortical areas [3]. Thus we observe a significant top-down feedback system, which becomes an important issue to consider when modeling the visual system. Figure 2: The geniculostriate pathway from the retina to the primary visual cortex in humans. Adapted from [5]. Neurons in each of the topographic maps interact in two ways: excitatory and inhibitory. In addition to interactions between maps, there is also a certain amount of lateral interaction between neurons of the same map. This further adds to the complexity of modeling neurons in the cortical map, as there is a multitude of input/output relationships. The primary visual cortex, or V1, is the most important cortical region in the occipital lobe of the brain, since almost all signals from the optic nerve will pass through this region. Although there are many other cortical regions (i.e. V2-V6) utilized in visual processing, usually only V1 is considered when modeling due to the complexity of interconnections between the various cortical regions. Beyond the cortical regions are other areas of the brain where visual processing is performed, such as at the temporal and parietal lobes. Overall, however, V1 is central to the processing of visual information. IV. MODELING THE VISUAL CORTEX: TOPOGRAPHICA/LISSOM Figure 1: Structure of the human eye. Adapted from [4]. The most dominant pathway for the optical signal in humans is the geniculostriate pathway, which is illustrated in Figure 2. The major termination for the optic tract nerve in this pathway is at the lateral geniculate nucleus (LGN). This structure contains topographic maps of the visual field, which are essentially projections of the visual field onto a network of neurons. The term map is utilized since the organization of this biological neural network is similar to a roadmap, in that the spatial relations between points of light on the retinal surface are preserved. Due to this organization, each lateral geniculate neuron is associated with a receptive field similar to One of the most recent models of the visual system is the Topographica software package [6], [7]. This is also one of the few models that are intended to complement current models for the neuron (e.g. NEURON, GENESIS). By default, however, the software utilizes a simple neuron model. In fact, the fundamental unit or component of the model is a sheet of neurons, which consists of a two-dimensional continuous area of a finite number of neurons, rather than just individual neurons. The significance of the Topographica model is that current simulators lack specific support for biologically realistic, densely interconnected topographic maps, as well as for generating input patterns at the topographic map level [7]. Using today’s more powerful computers, Topographica is capable of simulating networks of tens of thousands of 3 neurons, resulting in interconnections on the order of millions to tens of millions [7], which was not previously possible. rule, normalized such that the sum of weights from each type of RF ρ is constant for each neuron (i,j) [8]. The iterative nature of this calculation is due to the weights adapting as a function of previous weight values, as well as the current synapse properties: wij , ab( f 1) wij , ab( f ) ijX ab ab[ wij , ab( f ) ijX ab] (3) where ηij now represents the final activity for neuron (i,j), wij,ρab(f) is the connection weight from the previous fixation, α is the learning rate associated with each type of connection, and Xρab is the presynaptic activity. Overall, these equations demonstrate that neuronal weight change is determined by many factors, primarily the product of the pre- and post-synaptic activity, distances between laterally correlated neurons, as well as a learning rate [8]. V. EVALUATION OF THE MODEL Figure 3: Topographica model [6], [7] The underlying model in the Topographica software is the LISSOM model. The HLISSOM model is a version of LISSOM that also includes simulation of the LGN structure. The following demonstrates the process that occurs in this model via the respective equations at each stage of Figure 3. For more details on the equations, refer to [8]. Firstly, the input to the model consists of the activity patterns on the sheet of photoreceptors, such as grayscale images. The response of each LGN unit (i,j) is then calculated as a scalar product of a fixed weight vector and its receptive field (RF) on the photoreceptor sheet [8]: ij ( ab X abwij , ab) (1) where σ is a piecewise linear sigmoid activation function, Xρab is the activation of input unit (a,b) in RF ρ, wij,ρab is the corresponding weight value, and γρ is a constant scaling factor. [8] The V1 neurons are calculated in a similar fashion. Iterations of the formula are required, however, since the V1 activity settles after the effects of short-range excitatory and long-range inhibitory lateral interaction. The variable s illustrates the iterative nature of this calculation, since Xρab(s-1) is the activation of input (a,b) during the previous settling step. Furthermore, ρ takes two values, corresponding to the RF’s on ON and OFF LGN sheets [8]. ij ( s) ( ab X ab( s 1) wij , ab) Models serve to simplify the complexities of reality by making assumptions. These assumptions can be general principles or specific simplifications in the experiment. Although this allows modelers to reduce the dimensions of a complex problem, difficulties can arise in neurobiology, since an assumption may ease development of a physical model, but have no application in biology. In other words, there is no guarantee that biological mechanisms will adhere to a modeler’s assumptions [9]. The goal in evaluating a model, therefore, is to conclude whether or not the assumptions made are supported by experimental observations. However, this does not necessarily validate the model in its entirety, since the collaboration of various assumptions can imply other assumptions that are not necessarily well tested in biology. Most models of the visual cortex, including the Topographica model, have a common set of assumptions [9]: (2) Once the activity of the V1 neuron has settled (i.e. after (2) has converged), the V1 weights adapt according to the Hebb Hebb synapses correlated or spatially patterned activity in the afferents to cortical neurons fixed connections between cortical neurons which are locally excitatory and inhibitory at slightly greater distances normalization of synapse strength Certain types of simplifications can lead to powerful and elegant models, but can become difficult to relate to the actual biological systems. At the same time, an extreme amount of detail in the model can increase the possibility of disproving the model, and thus components must be carefully chosen according to the desired functionality [9]. For the Topographica model, evaluation consisted of simulating a two-stage process of development, based on prenatal and postnatal training. By doing this, it was shown that the model network first develops an initial orientation map 4 through prenatal spontaneous activity, then gradually refines based on experience with postnatal natural images, without changing the overall shape of the map. This has been supported by data collected in the ferret visual cortex [8]. The significance of this testing methodology is that training and test data (i.e. images) can be closely related to tests done in biology. However, since this model only interfaces with an external environment, image data must be carefully chosen in order to conclusively correlate predicted results with experimental results. This is the fundamental requirement in Steps 2 and 3 of the “Framework of Systems Biology”. The basic question that comes out of this analysis is the validity of assumptions. In the case of a Hebb-based model such as Topographica, it must be investigated whether or not it is sufficient to consider this strict local learning rule, in which only pre- and postsynaptic elements are involved [10]. It may be possible that non-local effects should be incorporated into the synapse. Another more general issue is that of the actual computational processes taking place within the cortical maps [10]. In the case of the visual cortex, biological data explaining the internal connections within each cortical area is relatively low, with the exception of the primary visual cortex, which has been more extensively studied and thus is the focus of modern visual system models. VI. COMPLEXITY IN MODELING In addition to the issue of creating assumptions in models, the problem of dealing with complexity is significant, as it essentially affects all systems biology-based modeling. A high amount of feedback has been physiologically observed in the visual system. In systems biology this has profound effects on the difficulty of modeling. Consider the ability of a control systems engineer, who is capable of performing open-loop experiments that isolate forward and reverse pathways in a system by removing feedback. Compare this to a biological system, such as the visual cortex, which contains multiple-input multiple-output systems, and high amounts of interconnected forward and reverse pathways that simply cannot be isolated. This is a fundamental issue in systems biology in general, and is exemplified by the visual cortex example. Complexity in such biological networks is largely due to the adaptive nature of biological systems. Evolution has shown that survival is largely based on robustness, and thus most biological systems are naturally inclined to contain high amounts of redundancy. This relates back to the issue of assumptions, since modelers must be careful not to over-simplify model structure and dynamics. Details that are difficult to measure within a biological system may have a large effect on overall inputoutput relationships, and so the model plan must be capable of accommodating such complexities. VII. CONCLUSION Despite the various issues involved in evaluating visual cortex models, the Topographica model provides a good example of the systems biology approach. A key attribute of this model is that it fills and important gap between existing detailed models of individual neurons and higher-level models for cognitive processes [7]. Thus the components of the model are well defined and adaptable to new discoveries or requirements at a fundamental (i.e. neuronal) level. Furthermore, the functionality of the model is designed in a hierarchical fashion, such that further refinements can be made based on new experiments. This iterative design is vital in the systems biology approach. Issues exist, however, when considering the accuracy of the model as well as the inherent assumptions. Although many aspects of map formation have been measured and modeled in great detail, many functional questions remain. Future work in this field, therefore, should consist of conclusively validating the functionality of visual cortex models with actual biological data. REFERENCES T. Ideker, T. Galitski, and L. Hood, “A new approach to decoding life: Systems biology,” Annu. Rev. Genomics. Hum. Genet., vol. 2, pp. 343372, 2001. [2] H. Kitano, “Looking beyond the details: A rise in system-oriented approaches in genetics and molecular biology,” Curr. Genet., vol. 41, pp. 1-10, 2002. [3] S. Coren, L. M. Ward, and J. T. Enns, Sensation and Perception (5th ed.), Harcourt College Publishers, 1999, pp. 50-84. [4] URL: http://www.optom.demon.co.uk/elderly/fig1.gif [5] URL: http://www.fz-juelich.de/ibi/ibi-1/datapool/page/181/figure%205500.jpg [6] Neural Networks Research Group, University of Texas at Austin, “The Topographica simulator”, October 2004, Available: http://topographica.org [7] J. A. Bednar et al., “Modeling cortical maps with Topographica”, Neurocomputing, in press. Presented at the 2003 Computational Neuroscience meeting, 2004. [8] J. A. Bednar, and R. Miikkulainen, “Prenatal and postnatal development of laterally connected orientation maps”, Neurocomputing, in press. Presented at the 2003 Computational Neuroscience meeting, 2004. [9] N. V. Swindale, “The development of topography in the visual cortex: a review of models”, Network: Comput. Neural Syst., vol. 7, pp. 161-247, 1996. [10] H. Preibl, “Cortical maps”, in Information Processing in the Cortex, A. Aertsen, V. Braitenberg, Eds., Berlin: Springer-Verlag, 1992, pp. 451457 [1]