* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download NUCLEOTIDE METABOLISM
Polyadenylation wikipedia , lookup
Lipid signaling wikipedia , lookup
Proteolysis wikipedia , lookup
Point mutation wikipedia , lookup
Genetic code wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Butyric acid wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Peptide synthesis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Citric acid cycle wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Biochemistry wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
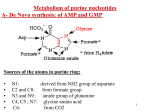
NUCLEOTIDE METABOLISM SITI ANNISA DEVI TRUSDA Nucleotides are essential for all cells DNA/RNA synthesisprotein synthesiscells proliferate Carriers of activated intermediates in the synthesis of carbohydrate, lipids and protein Structural component of several essential coenzymes (coA,FAD,NAD+,NADP+) cAMP,cGMP2nd messenger in signal transduction pathway Important regulatory compound for many of the pathways of intermediary metabolism, inhibiting/activating key enzimes Nucleotide structure Consist of: ◦ Nitrogenous base : purine & pyrimidine ◦ Pentose monosaccharide ◦ 1/2/3 phosphate groups DNA and RNA contain the same purine bases: A & G Pirimidine RNA : U & C DNA : T & C T& U differ by only one methyl group Nucleosides Pentose sugar + Nitrogen Base = Nucleosides So, nucleotides = Nucleosides + Phosphate If the sugar is ribose : ribonucleosides If deoxyribose: deoxyribonucleosides Ribonucleosides of A,G,C,U: Adenosine,Guanosine,Cytidine,Uridine What are the deoxyribonucleosides for A,G,C,T? Nucleotides mono,di,tri esters of nucleosides 1st phosphate group is attached by an ester linkage to the 5’OH of the pentosenucleoside 5’phosphate/5’-nucleoside Type of pentose is added as prefix for nucleotide, can be ribose/deoxyribose e.g: 5’ribonucleotide/5’-deoxyribonucleotide 1 phosphate group + 5’-carbon of the pentosenucleoside monophosphate(NMP) e.g AMP, CMP 2 or 3 phosphate group added to the nucleosidenucleoside di/triphosphate e.g ADP/ATP The latter connected to the nucleotide by a highenergy bond Phosphate groups(-) charge DNA/RNA=nucleic acids So, what is : ◦ Nucleoside? ◦ Nucleotide? ◦ Nucleic acid? SYNTHESIS OF PURINE NUCLEOTIDES Source of purine ring: aspartic acid, glycine, glutamine, CO2,N10-formylTHF Synthesis of 5-phosphoribosyl-1pyrophosphate (PRPP) an activated pentose for synthesis of purine/pirimidine & salvage of purine bases catalyzed by PRPP synthetase, from ATP & ribose 5-phosphate this enzyme is activated by inorganic phosphat (Pi), inhibited by purine nucleotides the sugar of PRPP is ribose ribonucleotides as end product of purine synthetis Purine synthesis is critical to fetal development, therefore defects in enzymes will result in a nonviable fetus. PRPP synthetase defects are known and have severe consequences (next slide) PRPP synthetase superactivity has been documented, resulting in increased PRPP, elevated levels of nucleotides, and increased excretion of uric acid. Phosphoribosyl Pyrophosphate (PRPP) Synthetase Defects PRPP deficiency results in convulsions, autistic behavior, anemia, and severe mental retardation. Excessive PRPP activity causes gout (deposition of uric acid crystals), along with various neurological symptoms, such as deafness. Synthesis of 5’-phosphoribosylamine Amide group of glutamine replaces the pyrophosphate group at C1 of PRPP the enzyme, glutamine:phosphoribosyl pyrophosphate amidotransferase is inhibited by the purine 5’-nucleotides AMP,GMP,IMP (end product) Committed step Rate of reaction also controlled by intracellular [] of glutamine and PRPP Synthesis of inosine monophosphate,the “parent” of purine nucleotide requires 4 ATP 2 steps require N10 –formyltetrahydrofolate Conversion of IMP to AMP and GMP 2 step energy requiring pathway synthesis of AMP requires GTP as energy source synthesis of GMP requires ATP Conversion of nucleoside monophosphates to nucleoside di and triphosphate AMP + ATP ↔ 2 ADP GMP +ATP ↔ GDP + ADP GDP + ATP ↔ GTP + ADP CDP + ATP ↔ CTP + ADP Purine Synthesis Adenilosuksinat synthetase IMP dehidrogenase XMP aminase Adenilosuksinat lyase DAUR dr IMP AMP & GMP Salvage Pathway of purines Purines that result from the normal turnover of cellular nucleic acids/diet can be reconverted into nucleoside triphosphatessalvage pathway 2 enzymes: Adenine phosphoribosyltransferase (APRT), and hypoxanthine-guanine phosphoribosyltransferase (HPRT) Both needs PRPP as the source of the ribose 5-phosphate Degradation of Purine Nucleotides Purine Nucleotides from ingested nucleic acids or turnover of cellular nucleic acids is excreted by humans as uric acid. Humans excrete about 0.6 g uric acid every 24 hours. Degradation of dietary nucleic acids occurs in the small intestine by pancreatic enzymes Digestion of dietary nucleic acids In the stomach: low pH denatures DNA&RNA In small intestine: break down phosphodiester bond by endonuclease (pancreas) oligonucleotide By phosphodiesterase(exonuclease non spesific enzyme) mononucleotide By phosphomonoesterase (nucleotidase) result: nucleoside and orthophosphate. Nucleosida phosphorylase result: base and ribose-1-phosphate. The nucleoside then absorbed by intestinal mucosal cells If the base or nucleoside is unused, it will be reused in salvage pathways, the base will be degraded: uric acid (purin) ureidopropionic (pyrimidine). Diseases associated with purine degradation Gout Elevated uric acid levels in the blood Uric acid crystals will form in the extremities with a surrounding area of inflammation. This is called a tophus and is often described as an arthritic “great toe”. Can be caused by a defect in an enzyme of purine metabolism or by reduced secretion of uric acid into the urinary tract. tophus Adenosine Deaminase (ADA) and Purine Nucleoside Phosphorylase (PNP) Deficiency. accumulation of adenosine wich is converted to its ribonucleotide or deoxyribonucleotide form by cellular kinases As dATP level rise, ribonucleotide reductase is inhibited↓ production of all deoxyribose containing nucleotidescells cannot make DNA and divide. Most severe form: severe combined immunodeficiency disease (SCID)lack of T and B cells A deficiency of either ADA or PNP causes a moderate to complete lack of immune function. Affected children cannot survive outside a sterile environment. They may also have moderate neurological problems, including partial paralysis of the limbs. When a compatible donor can be found, bone marrow transplant is an effective treatment. Lesch-Nyhan Syndrome Hypoxanthine Guanine Phosphoribosyltransferase (HGPRT) deficiency X-linked genetic condition Severe neurologic disease, characterized by selfmutilating behaviors such as lip and finger biting and/or head banging Up to 20 times the uric acid in the urine than in normal individuals. Uric acid crystals form in the urine. Untreated condition results in death within the first year due to kidney failure. Treated with allopurinol, a competitive inhibitor of xanthine oxidase. SYNTHESIS OF DEOXYRIBONUCLEOTIDES Deoxyribonucleotides required for DNA synthesis (2’-deoxyribonucleotides) Enzyme: ribonucleotide reductase Inhibitor : dATP Needed a coenzyme : thioredoxin Thioredoxin is regenerated by thioredoxin reductase Regulation of ribonucleotide reduction is controlled by allosteric feedback mechanisms. PYRIMIDINE SYNTHESIS AND DEGRADATION Source of pyrimidine ring: glutamine, CO2, aspartic acid Synthesis of carbamoyl phosphate from glutamine & CO2, enzyme: carbamoyl phosphate synthetase II (CPS II), inhibited by UTP activated by ATP and PRPP Synthesis of orotic acid formation of carbamoylaspartatedihydroorotateorotic acid (mind the enzymes!!) Formation of a pyrimidine nucleotide : orotidine 5’-monophosphate (OMP)the parent of pyrimidine mononucleotide OMPUridine monophosphate (UMP) Synthesis of uridine triphosphate and cytidine triphosphate CTP is produced by amination of UTP Synthesis of thymidine monophosphate from dUMP Orotat fosforibosiltransferase CTP synthetase Orotidilate dekarboksilase UMP kinase Nukleosida diphosphat kinase Pyrimidine Synthesis Production of Uridine 5’-monophosphate (UMP) from orotate is catalyzed by the enzyme UMP synthase Orotic Aciduria Deficiency in UMP synthetase activity Due to the demand for nucleotides in the process of red blood cell synthesis, patients develop the condition of megaloblastic anemia, a deficiency of red blood cells. Pyrimidine synthesis is decreased and excess orotic acid is excreted in the urine (hence the name orotic aciduria) Degradation of pyrimidine nucleotides Unlike the purine rings, which are not cleaved in human cells, the pyrimidine ring can be opened and degraded to highly soluble structures, such as β-alanine, and β-aminoisobutyrate, which can serve as precursors of acetyl coA and succinyl coA SALVAGE OF PYRIMIDINES Pyrimidine salvage defects have not been clinically documented